Validation of a redesigned pan-poliovirus assay and real-time PCR platforms for the global poliovirus laboratory network
- PMID: 34358268
- PMCID: PMC8345876
- DOI: 10.1371/journal.pone.0255795
Validation of a redesigned pan-poliovirus assay and real-time PCR platforms for the global poliovirus laboratory network
Erratum in
-
Correction: Validation of a redesigned pan-poliovirus assay and real-time PCR platforms for the global poliovirus laboratory network.PLoS One. 2024 Aug 2;19(8):e0308467. doi: 10.1371/journal.pone.0308467. eCollection 2024. PLoS One. 2024. PMID: 39093885 Free PMC article.
Abstract
Surveillance and detection of polioviruses (PV) remain crucial to monitoring eradication progress. Intratypic differentiation (ITD) using the real-time RT-PCR kit is key to the surveillance workflow, where viruses are screened after cell culture isolation before a subset are verified by sequencing. The ITD kit is a series of real-time RT-PCR assays that screens cytopathic effect (CPE)-positive cell cultures using the standard WHO method for virus isolation. Because ITD screening is a critical procedure in the poliovirus identification workflow, validation of performance of real-time PCR platforms is a core requirement for the detection of poliovirus using the ITD kit. In addition, the continual update and improvement of the ITD assays to simplify interpretation in all platforms is necessary to ensure that all real-time machines are capable of detecting positive real-time signals. Four platforms (ABI7500 real-time systems, Bio-Rad CFX96, Stratagene MX3000P, and the Qiagen Rotor-Gene Q) were validated with the ITD kit and a redesigned poliovirus probe. The poliovirus probe in the real-time RT-PCR pan-poliovirus (PanPV) assay was re-designed with a double-quencher (Zen™) to reduce background fluorescence and potential false negatives. The updated PanPV probe was evaluated with a panel consisting of 184 polioviruses and non-polio enteroviruses. To further validate the updated PanPV probe, the new assay was pilot tested in five Global Polio Laboratory Network (GPLN) laboratories (Madagascar, India, Philippines, Pakistan, and Democratic Republic of Congo). The updated PanPV probe performance was shown to reduce background fluorescence and decrease the number of false positives compared to the standard PanPV probe.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
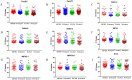


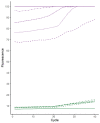

Similar articles
-
Culture-Independent Detection of Poliovirus in Stool Samples by Direct RNA Extraction.Microbiol Spectr. 2021 Dec 22;9(3):e0066821. doi: 10.1128/Spectrum.00668-21. Epub 2021 Nov 10. Microbiol Spectr. 2021. PMID: 34756088 Free PMC article.
-
Advancing poliovirus eradication: lessons learned from piloting direct molecular detection of polioviruses in high-risk and priority geographies.Microbiol Spectr. 2025 Feb 4;13(2):e0227924. doi: 10.1128/spectrum.02279-24. Epub 2024 Dec 12. Microbiol Spectr. 2025. PMID: 39665559 Free PMC article.
-
Diagnostic Assay Development for Poliovirus Eradication.J Clin Microbiol. 2018 Jan 24;56(2):e01624-17. doi: 10.1128/JCM.01624-17. Print 2018 Feb. J Clin Microbiol. 2018. PMID: 29212703 Free PMC article.
-
Circulating vaccine-derived polioviruses: current state of knowledge.Bull World Health Organ. 2004 Jan;82(1):16-23. Epub 2004 Feb 26. Bull World Health Organ. 2004. PMID: 15106296 Free PMC article. Review.
-
Wild and vaccine-derived poliovirus circulation, and implications for polio eradication.Epidemiol Infect. 2017 Feb;145(3):413-419. doi: 10.1017/S0950268816002569. Epub 2016 Nov 21. Epidemiol Infect. 2017. PMID: 27866483 Free PMC article. Review.
Cited by
-
Characterization of environmental and clinical surveillance inputs to support prospective integrated modeling of the polio endgame.PLOS Glob Public Health. 2025 Feb 7;5(2):e0004168. doi: 10.1371/journal.pgph.0004168. eCollection 2025. PLOS Glob Public Health. 2025. PMID: 39919149 Free PMC article. Review.
-
Wastewater Testing and Detection of Poliovirus Type 2 Genetically Linked to Virus Isolated from a Paralytic Polio Case - New York, March 9-October 11, 2022.MMWR Morb Mortal Wkly Rep. 2022 Nov 4;71(44):1418-1424. doi: 10.15585/mmwr.mm7144e2. MMWR Morb Mortal Wkly Rep. 2022. PMID: 36327157 Free PMC article.
-
Evaluation of Direct Detection Protocols for Poliovirus from Stool Samples of Acute Flaccid Paralysis Patients.Viruses. 2023 Oct 18;15(10):2113. doi: 10.3390/v15102113. Viruses. 2023. PMID: 37896890 Free PMC article.
-
Comparison of Eleven RNA Extraction Methods for Poliovirus Direct Molecular Detection in Stool Samples.Microbiol Spectr. 2023 Mar 20;11(2):e0425222. doi: 10.1128/spectrum.04252-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36939356 Free PMC article.
-
A comparative study on environmental surveillance of enterovirus: Using a two-phase separation method and a filtration method with a mixed cellulose ester (MCE) membrane.Biosaf Health. 2023 Apr 11;5(3):174-180. doi: 10.1016/j.bsheal.2023.04.001. eCollection 2023 Jun. Biosaf Health. 2023. PMID: 40078513 Free PMC article.
References
-
- WHO. Expanded programme on immunization. Progress towards the global eradication of poliomyelitis, 1995. Wkly Epidemiol Rec. 1996;71(25):189–94. Epub 1996/06/21. . - PubMed
-
- Kew OM, Mulders MN, Lipskaya GY, da Silva EE, Patlansch MA. Molecular epidemiology of polioviruses. Seminars in Virology. 1995;6(6):401–14. doi: 10.1016/S1044-5773(05)80017-4 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

