Clinical experience with non-invasive prenatal screening for single-gene disorders
- PMID: 34358384
- PMCID: PMC9302116
- DOI: 10.1002/uog.23756
Clinical experience with non-invasive prenatal screening for single-gene disorders
Abstract
Objective: To assess the performance of a non-invasive prenatal screening test (NIPT) for a panel of dominant single-gene disorders (SGD) with a combined population incidence of 1 in 600.
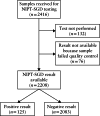
Methods: Cell-free fetal DNA isolated from maternal plasma samples accessioned from 14 April 2017 to 27 November 2019 was analyzed by next-generation sequencing, targeting 30 genes, to look for pathogenic or likely pathogenic variants implicated in 25 dominant conditions. The conditions included Noonan spectrum disorders, skeletal disorders, craniosynostosis syndromes, Cornelia de Lange syndrome, Alagille syndrome, tuberous sclerosis, epileptic encephalopathy, SYNGAP1-related intellectual disability, CHARGE syndrome, Sotos syndrome and Rett syndrome. NIPT-SGD was made available as a clinical service to women with a singleton pregnancy at ≥ 9 weeks' gestation, with testing on maternal and paternal genomic DNA to assist in interpretation. A minimum of 4.5% fetal fraction was required for test interpretation. Variants identified in the mother were deemed inconclusive with respect to fetal carrier status. Confirmatory prenatal or postnatal diagnostic testing was recommended for all screen-positive patients and follow-up information was requested. The screen-positive rates with respect to the clinical indication for testing were evaluated.
Results: A NIPT-SGD result was available for 2208 women, of which 125 (5.7%) were positive. Elevated test-positive rates were observed for referrals with a family history of a disorder on the panel (20/132 (15.2%)) or a primary indication of fetal long-bone abnormality (60/178 (33.7%)), fetal craniofacial abnormality (6/21 (28.6%)), fetal lymphatic abnormality (20/150 (13.3%)) or major fetal cardiac defect (4/31 (12.9%)). For paternal age ≥ 40 years as a sole risk factor, the test-positive rate was 2/912 (0.2%). Of the 125 positive cases, follow-up information was available for 67 (53.6%), with none classified as false-positive. No false-negative cases were identified.
Conclusions: NIPT can assist in the early detection of a set of SGD, particularly when either abnormal ultrasound findings or a family history is present. Additional clinical studies are needed to evaluate the optimal design of the gene panel, define target populations and assess patient acceptability. NIPT-SGD offers a safe and early prenatal screening option. © 2021 The Authors. Ultrasound in Obstetrics & Gynecology published by John Wiley & Sons Ltd on behalf of International Society of Ultrasound in Obstetrics and Gynecology.
Keywords: Noonan spectrum disorder; autosomal dominant disorder; cell-free DNA; next-generation sequencing; non-invasive prenatal testing; single-gene disorder.
© 2021 The Authors. Ultrasound in Obstetrics & Gynecology published by John Wiley & Sons Ltd on behalf of International Society of Ultrasound in Obstetrics and Gynecology.
Figures
References
-
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet 1997; 350: 485–487. - PubMed
-
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Deciu C, Grody WW, Nelson SF, Canick JA. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med 2011; 13: 913–920. - PubMed
-
- Palomaki GE, Deciu C, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Grody WW, Nelson SF, Canick JA. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med 2012; 14: 296–305. - PMC - PubMed
-
- Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP. Genome‐wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol 2012; 119: 890–901. - PubMed
-
- Pergament E, Cuckle H, Zimmermann B, Banjevic M, Sigurjonsson S, Ryan A, Hall MP, Dodd M, Lacroute P, Stosic M, Chopra N, Hunkapiller N, Prosen DE, McAdoo S, Demko Z, Siddiqui A, Hill M, Rabinowitz M. Single‐nucleotide polymorphism‐based noninvasive prenatal screening in a high‐risk and low‐risk cohort. Obstet Gynecol 2014; 124: 210–218. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous