Cell-type-specific Hox regulatory strategies orchestrate tissue identity
- PMID: 34358443
- PMCID: PMC8511240
- DOI: 10.1016/j.cub.2021.07.030
Cell-type-specific Hox regulatory strategies orchestrate tissue identity
Abstract
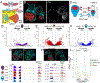
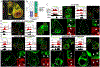
Hox proteins are homeodomain transcription factors that diversify serially homologous segments along the animal body axis, as revealed by the classic bithorax phenotype of Drosophila melanogaster, in which mutations in Ultrabithorax (Ubx) transform the third thoracic segment into the likeness of the second thoracic segment. To specify segment identity, we show that Ubx both increases and decreases chromatin accessibility, coinciding with its dual role as both an activator and repressor of transcription. However, the choice of transcriptional activity executed by Ubx is spatially regulated and depends on the availability of cofactors, with Ubx acting as a repressor in some populations and as an activator in others. Ubx-mediated changes to chromatin accessibility positively and negatively affect the binding of Scalloped (Sd), a transcription factor that is required for appendage development in both segments. These findings illustrate how a single Hox protein can modify complex gene regulatory networks to transform the identity of an entire tissue.
Keywords: Drosophila melanogaster; Hox cofactors; Hox genes; Ubx; Ultrabithorax; appendage development; bithorax; haltere; serial homology; wing.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
Gene regulation: Context is everything.Curr Biol. 2021 Oct 11;31(19):R1115-R1117. doi: 10.1016/j.cub.2021.08.064. Curr Biol. 2021. PMID: 34637709
References
-
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978. December;276(5688):565–70. - PubMed
-
- Bridges CB The mutants of Drosophila melanogaster. Vol. 552. Carnegie Inst. Wash. Publ.; 1944.
-
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, et al. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983. July 1;221(4605):23–9. - PubMed
-
- Ghosh P, Sagerström CG. Developing roles for Hox proteins in hindbrain gene regulatory networks. Int J Dev Biol. 2018;62(11–12):767–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

