Quantitative Assessment and Prognostic Associations of the Immune Landscape in Ovarian Clear Cell Carcinoma
- PMID: 34359755
- PMCID: PMC8345766
- DOI: 10.3390/cancers13153854
Quantitative Assessment and Prognostic Associations of the Immune Landscape in Ovarian Clear Cell Carcinoma
Abstract
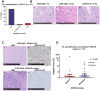
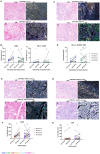
Ovarian clear cell carcinoma (OCCC) is a rare subtype of epithelial ovarian cancer characterised by a high frequency of loss-of-function ARID1A mutations and a poor response to chemotherapy. Despite their generally low mutational burden, an intratumoural T cell response has been reported in a subset of OCCC, with ARID1A purported to be a biomarker for the response to the immune checkpoint blockade independent of micro-satellite instability (MSI). However, assessment of the different immune cell types and spatial distribution specifically within OCCC patients has not been described to date. Here, we characterised the immune landscape of OCCC by profiling a cohort of 33 microsatellite stable OCCCs at the genomic, gene expression and histological level using targeted sequencing, gene expression profiling using the NanoString targeted immune panel, and multiplex immunofluorescence to assess the spatial distribution and abundance of immune cell populations at the protein level. Analysis of these tumours and subsequent independent validation identified an immune-related gene expression signature associated with risk of recurrence of OCCC. Whilst histological quantification of tumour-infiltrating lymphocytes (TIL, Salgado scoring) showed no association with the risk of recurrence or ARID1A mutational status, the characterisation of TILs via multiplexed immunofluorescence identified spatial differences in immunosuppressive cell populations in OCCC. Tumour-associated macrophages (TAM) and regulatory T cells were excluded from the vicinity of tumour cells in low-risk patients, suggesting that high-risk patients have a more immunosuppressive microenvironment. We also found that TAMs and cytotoxic T cells were also excluded from the vicinity of tumour cells in ARID1A-mutated OCCCs compared to ARID1A wild-type tumours, suggesting that the exclusion of these immune effectors could determine the host response of ARID1A-mutant OCCCs to therapy. Overall, our study has provided new insights into the immune landscape and prognostic associations in OCCC and suggest that tailored immunotherapeutic approaches may be warranted for different subgroups of OCCC patients.
Keywords: ARID1A; biomarker; clear cell ovarian cancer; immune microenvironment; next generation sequencing.
Conflict of interest statement
C.J.L. reports grants from CRUK and Breast Cancer Now during the conduct of the study, is a named inventor on patents describing the use of DNA repair inhibitors and stands to gain from their use as part of the Institute of Cancer Research “Rewards to Inventor” scheme, has received research funding from AstraZeneca, Merck KGaA, Artios, Pfizer, and consultancy and/or advisory fees from Artios, AstraZeneca, MerckKGaA, Tango, and GLG and is a shareholder of OviBio and Tango outside the submitted work. S.B. has worked in an advisory role for Amgen, AstraZeneca, Clovis Oncology, Epsilogen, Genmab, GlaxoSmithKline, Immunogen, Mersana, Merck Sharp & Dohme, Merck Sereno, Oncxerna, Pfizer, Tesaro and Roche, has received institution research funding from AstraZeneca and has received funding support from NIHR RM/ICR Biomedical Research Centre for Cancer. The remining authors declare no conflicts of interest.
Figures


References
-
- Tan D.S., Iravani M., McCluggage W.G., Lambros M.B., Milanezi F., Mackay A., Gourley C., Geyer F.C., Vatcheva R., Millar J., et al. Genomic analysis reveals the molecular heterogeneity of ovarian clear cell carcinomas. Clin. Cancer Res. 2011;17:1521–1534. doi: 10.1158/1078-0432.CCR-10-1688. - DOI - PubMed
-
- Goff B.A., de la Cuesta R.S., Muntz H.G., Fleischhacker D., Ek M., Rice L.W., Nikrui N., Tamimi H.K., Cain J.M., Greer B.E., et al. Clear cell carcinoma of the ovary: A distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy in stage III disease. Gynecol. Oncol. 1996;60:412–417. doi: 10.1006/gyno.1996.0065. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

