How Influenza Virus Uses Host Cell Pathways during Uncoating
- PMID: 34359892
- PMCID: PMC8305448
- DOI: 10.3390/cells10071722
How Influenza Virus Uses Host Cell Pathways during Uncoating
Abstract
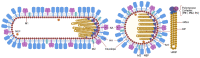
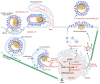
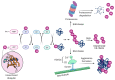
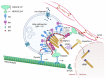
Influenza is a zoonotic respiratory disease of major public health interest due to its pandemic potential, and a threat to animals and the human population. The influenza A virus genome consists of eight single-stranded RNA segments sequestered within a protein capsid and a lipid bilayer envelope. During host cell entry, cellular cues contribute to viral conformational changes that promote critical events such as fusion with late endosomes, capsid uncoating and viral genome release into the cytosol. In this focused review, we concisely describe the virus infection cycle and highlight the recent findings of host cell pathways and cytosolic proteins that assist influenza uncoating during host cell entry.
Keywords: EPS8; HDAC6; M1; TNPO1; capsid uncoating; influenza; pandemic; ubiquitin; virus–host interaction.
Conflict of interest statement
The authors declare the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Burrell C.J., Howard C.R., Murphy F.A. Emerging Virus Diseases. Fenner White’s Med. Virol. 2017 doi: 10.1016/B978-0-12-375156-0.00015-1. - DOI
-
- Ryu W.-S. Chapter 21—New Emerging Viruses. In: Ryu W.-S., editor. Molecular Virology of Human Pathogenic Viruses. Academic Press; Boston, MA, USA: 2017. pp. 289–302. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

