TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions
- PMID: 34359920
- PMCID: PMC8303332
- DOI: 10.3390/cells10071750
TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions
Abstract
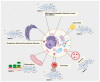
Transmembrane proteins (TMEMs) are integral proteins that span biological membranes. TMEMs function as cellular membrane gates by modifying their conformation to control the influx and efflux of signals and molecules. TMEMs also reside in and interact with the membranes of various intracellular organelles. Despite much knowledge about the biological importance of TMEMs, their role in metabolic regulation is poorly understood. This review highlights the role of a single TMEM, transmembrane protein 135 (TMEM135). TMEM135 is thought to regulate the balance between mitochondrial fusion and fission and plays a role in regulating lipid droplet formation/tethering, fatty acid metabolism, and peroxisomal function. This review highlights our current understanding of the various roles of TMEM135 in cellular processes, organelle function, calcium dynamics, and metabolism.
Keywords: TMEM135; aging; fission; mitochondrial dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Alberts B., editor; Alberts B., editor. Molecular Biology of the Cell. 5th ed. Garland Science; New York, NY, USA: 2008.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

