Two-Pore Channels Regulate Expression of Various Receptors and Their Pathway-Related Proteins in Multiple Ways
- PMID: 34359976
- PMCID: PMC8307395
- DOI: 10.3390/cells10071807
Two-Pore Channels Regulate Expression of Various Receptors and Their Pathway-Related Proteins in Multiple Ways
Abstract
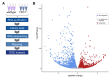
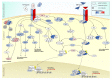
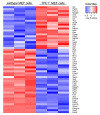
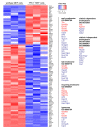
Two-pore channels (TPCs) constitute a small family of ion channels within membranes of intracellular acidic compartments, such as endosomes and lysosomes. They were shown to provide transient and locally restricted Ca2+-currents, likely responsible for fusion and/or fission events of endolysosomal membranes and thereby for intracellular vesicle trafficking. Genetic deletion of TPCs not only affects endocytosis, recycling, and degradation of various surface receptors but also uptake and impact of bacterial protein toxins and entry and intracellular processing of some types of viruses. This review points to important examples of these trafficking defects on one part but mainly focuses on the resulting impact of the TPC inactivation on receptor expression and receptor signaling. Thus, a detailed RNA sequencing analysis using TPC1-deficient fibroblasts uncovered a multitude of changes in the expression levels of surface receptors and their pathway-related signaling proteins. We refer to several classes of receptors such as EGF, TGF, and insulin as well as proteins involved in endocytosis.
Keywords: RNA sequence analysis; endolysosomal system; intracellular trafficking; receptor endocytosis; two-pore channel.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

