Human Mitochondrial RNA Processing and Modifications: Overview
- PMID: 34360765
- PMCID: PMC8348895
- DOI: 10.3390/ijms22157999
Human Mitochondrial RNA Processing and Modifications: Overview
Abstract
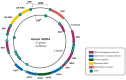
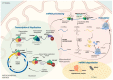
Mitochondria, often referred to as the powerhouses of cells, are vital organelles that are present in almost all eukaryotic organisms, including humans. They are the key energy suppliers as the site of adenosine triphosphate production, and are involved in apoptosis, calcium homeostasis, and regulation of the innate immune response. Abnormalities occurring in mitochondria, such as mitochondrial DNA (mtDNA) mutations and disturbances at any stage of mitochondrial RNA (mtRNA) processing and translation, usually lead to severe mitochondrial diseases. A fundamental line of investigation is to understand the processes that occur in these organelles and their physiological consequences. Despite substantial progress that has been made in the field of mtRNA processing and its regulation, many unknowns and controversies remain. The present review discusses the current state of knowledge of RNA processing in human mitochondria and sheds some light on the unresolved issues.
Keywords: RNA decay; RNA modifications; RNA processing; mitochondria; mitochondrial genome; mitochondrial transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

