Computational Analysis of the Crystal and Cryo-EM Structures of P-Loop Channels with Drugs
- PMID: 34360907
- PMCID: PMC8348670
- DOI: 10.3390/ijms22158143
Computational Analysis of the Crystal and Cryo-EM Structures of P-Loop Channels with Drugs
Abstract
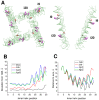
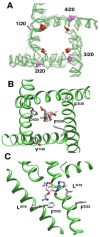
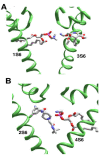
The superfamily of P-loop channels includes various potassium channels, voltage-gated sodium and calcium channels, transient receptor potential channels, and ionotropic glutamate receptors. Despite huge structural and functional diversity of the channels, their pore-forming domain has a conserved folding. In the past two decades, scores of atomic-scale structures of P-loop channels with medically important drugs in the inner pore have been published. High structural diversity of these complexes complicates the comparative analysis of these structures. Here we 3D-aligned structures of drug-bound P-loop channels, compared their geometric characteristics, and analyzed the energetics of ligand-channel interactions. In the superimposed structures drugs occupy most of the sterically available space in the inner pore and subunit/repeat interfaces. Cationic groups of some drugs occupy vacant binding sites of permeant ions in the inner pore and selectivity-filter region. Various electroneutral drugs, lipids, and detergent molecules are seen in the interfaces between subunits/repeats. In many structures the drugs strongly interact with lipid and detergent molecules, but physiological relevance of such interactions is unclear. Some eukaryotic sodium and calcium channels have state-dependent or drug-induced π-bulges in the inner helices, which would be difficult to predict. The drug-induced π-bulges may represent a novel mechanism of gating modulation.
Keywords: 3D alignment; Monte Carlo minimization; ligand-receptor interactions; sequence alignment; π-bulges.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
P-Loop Channels: Experimental Structures, and Physics-Based and Neural Networks-Based Models.Membranes (Basel). 2022 Feb 16;12(2):229. doi: 10.3390/membranes12020229. Membranes (Basel). 2022. PMID: 35207150 Free PMC article. Review.
-
Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure.Mol Pharmacol. 2012 Jul;82(1):97-104. doi: 10.1124/mol.112.078212. Epub 2012 Apr 13. Mol Pharmacol. 2012. PMID: 22505150
-
Computational Structural Pharmacology and Toxicology of Voltage-Gated Sodium Channels.Curr Top Membr. 2016;78:117-44. doi: 10.1016/bs.ctm.2015.12.001. Epub 2016 Mar 14. Curr Top Membr. 2016. PMID: 27586283
-
The pore domain in glutamate-gated ion channels: Structure, drug binding and similarity with potassium channels.Biochim Biophys Acta Biomembr. 2020 Oct 1;1862(10):183401. doi: 10.1016/j.bbamem.2020.183401. Epub 2020 Jun 17. Biochim Biophys Acta Biomembr. 2020. PMID: 32562696
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
Cited by
-
Modulation of Pore Opening of Eukaryotic Sodium Channels by π-Helices in S6.J Phys Chem Lett. 2023 Jun 29;14(25):5876-5881. doi: 10.1021/acs.jpclett.3c00803. Epub 2023 Jun 21. J Phys Chem Lett. 2023. PMID: 37341700 Free PMC article.
-
Mechanisms of dihydropyridine agonists and antagonists in view of cryo-EM structures of calcium and sodium channels.J Gen Physiol. 2023 Nov 6;155(11):e202313418. doi: 10.1085/jgp.202313418. Epub 2023 Sep 20. J Gen Physiol. 2023. PMID: 37728574 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

