Identification of an IL-22-Dependent Gene Signature as a Pharmacodynamic Biomarker
- PMID: 34360971
- PMCID: PMC8347589
- DOI: 10.3390/ijms22158205
Identification of an IL-22-Dependent Gene Signature as a Pharmacodynamic Biomarker
Abstract
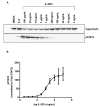
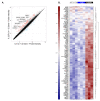
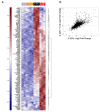
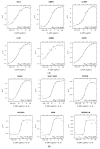
Interleukin-22 (IL-22) plays a role in epithelial barrier function and repair, and may provide benefits in conditions like inflammatory bowel disease. However, limited human data are available to assess the clinical effect of IL-22 administration. This study used a human intestinal cell line to identify an IL-22-dependent gene signature that could serve as a pharmacodynamic biomarker for IL-22 therapy. The response to IL-22Fc (UTTR1147A, an Fc-stabilized version of IL-22) was assessed in HT-29 cells by microarray, and the selected responsive genes were confirmed by qPCR. HT-29 cells demonstrated dose-dependent increases in STAT3 phosphorylation and multiple gene expression changes in response to UTTR1147A. Genes were selected that were upregulated by UTTR1147A, but to a lesser extent by IL-6, which also signals via STAT3. IL-1R1 was highly upregulated by UTTR1147A, and differential gene expression patterns were observed in response to IL-22Fc in the presence of IL-1β. An IL-22-dependent gene signature was identified that could serve as a pharmacodynamic biomarker in intestinal biopsies to support the clinical development of an IL-22 therapeutic. The differential gene expression pattern in the presence of IL-1β suggests that an inflammatory cytokine milieu in the disease setting could influence the clinical responses to IL-22.
Keywords: IBD; IL-1β; IL-22; antimicrobial; epithelial repair; gene expression; pharmacodynamic biomarker.
Conflict of interest statement
All of the authors are employees and stock holders of Genentech/Roche.
Figures

Similar articles
-
Pre-clinical and translational pharmacology of a human interleukin-22 IgG fusion protein for potential treatment of infectious or inflammatory diseases.Biochem Pharmacol. 2018 Jun;152:224-235. doi: 10.1016/j.bcp.2018.03.031. Epub 2018 Mar 31. Biochem Pharmacol. 2018. PMID: 29608910
-
Randomized Phase I Healthy Volunteer Study of UTTR1147A (IL-22Fc): A Potential Therapy for Epithelial Injury.Clin Pharmacol Ther. 2019 Jan;105(1):177-189. doi: 10.1002/cpt.1164. Epub 2018 Aug 12. Clin Pharmacol Ther. 2019. PMID: 29952004 Clinical Trial.
-
Nonclinical safety assessment of a human interleukin-22FC IG fusion protein demonstrates in vitro to in vivo and cross-species translatability.Pharmacol Res Perspect. 2018 Nov 15;6(6):e00434. doi: 10.1002/prp2.434. eCollection 2018 Dec. Pharmacol Res Perspect. 2018. PMID: 30464842 Free PMC article.
-
A role for interleukin-33 in T(H)2-polarized intestinal inflammation?Mucosal Immunol. 2011 Sep;4(5):496-502. doi: 10.1038/mi.2011.22. Epub 2011 May 25. Mucosal Immunol. 2011. PMID: 21613987 Review.
-
Interleukin-19 as an Immunoregulatory Cytokine.Curr Mol Pharmacol. 2021;14(2):191-199. doi: 10.2174/1874467213666200424151528. Curr Mol Pharmacol. 2021. PMID: 32329704 Review.
Cited by
-
Population pharmacokinetics and pharmacodynamics of efmarodocokin alfa (IL-22Fc).J Pharmacokinet Pharmacodyn. 2024 Apr;51(2):141-153. doi: 10.1007/s10928-023-09888-2. Epub 2023 Oct 20. J Pharmacokinet Pharmacodyn. 2024. PMID: 37864000
-
Hepatic vagotomy blunts liver regeneration after hepatectomy by downregulating the expression of interleukin-22.World J Gastrointest Surg. 2023 Dec 27;15(12):2866-2878. doi: 10.4240/wjgs.v15.i12.2866. World J Gastrointest Surg. 2023. PMID: 38222006 Free PMC article.
References
-
- Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.M., Diepolder H., Marquardt A., Jagla W., Popp A., et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

