Biocontrol of Biofilm Formation: Jamming of Sessile-Associated Rhizobial Communication by Rhodococcal Quorum-Quenching
- PMID: 34361010
- PMCID: PMC8347015
- DOI: 10.3390/ijms22158241
Biocontrol of Biofilm Formation: Jamming of Sessile-Associated Rhizobial Communication by Rhodococcal Quorum-Quenching
Abstract
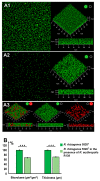
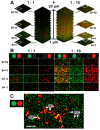
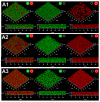
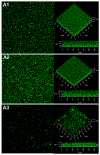
Biofilms are complex structures formed by a community of microbes adhering to a surface and/or to each other through the secretion of an adhesive and protective matrix. The establishment of these structures requires a coordination of action between microorganisms through powerful communication systems such as quorum-sensing. Therefore, auxiliary bacteria capable of interfering with these means of communication could be used to prevent biofilm formation and development. The phytopathogen Rhizobium rhizogenes, which causes hairy root disease and forms large biofilms in hydroponic crops, and the biocontrol agent Rhodococcus erythropolis R138 were used for this study. Changes in biofilm biovolume and structure, as well as interactions between rhizobia and rhodococci, were monitored by confocal laser scanning microscopy with appropriate fluorescent biosensors. We obtained direct visual evidence of an exchange of signals between rhizobia and the jamming of this communication by Rhodococcus within the biofilm. Signaling molecules were characterized as long chain (C14) N-acyl-homoserine lactones. The role of the Qsd quorum-quenching pathway in biofilm alteration was confirmed with an R. erythropolis mutant unable to produce the QsdA lactonase, and by expression of the qsdA gene in a heterologous host, Escherichia coli. Finally, Rhizobium biofilm formation was similarly inhibited by a purified extract of QsdA enzyme.
Keywords: N-acyl homoserine lactones; Rhizobium (Agrobacterium) rhizogenes; Rhodococcus erythropolis; biofilm; biological control; communication; hairy root; lifestyle switch; quorum-quenching; quorum-sensing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

