SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India
- PMID: 34361977
- PMCID: PMC8307577
- DOI: 10.3390/microorganisms9071542
SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India
Abstract
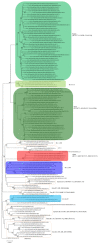
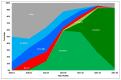
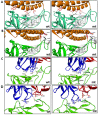
As the global severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic expands, genomic epidemiology and whole genome sequencing are being used to investigate its transmission and evolution. Against the backdrop of the global emergence of "variants of concern" (VOCs) during December 2020 and an upsurge in a state in the western part of India since January 2021, whole genome sequencing and analysis of spike protein mutations using sequence and structural approaches were undertaken to identify possible new variants and gauge the fitness of the current circulating strains. Phylogenetic analysis revealed that newly identified lineages B.1.617.1 and B.1.617.2 were predominantly circulating. The signature mutations possessed by these strains were L452R, T478K, E484Q, D614G and P681R in the spike protein, including within the receptor-binding domain (RBD). Of these, the mutations at residue positions 452, 484 and 681 have been reported in other globally circulating lineages. The structural analysis of RBD mutations L452R, T478K and E484Q revealed that these may possibly result in increased ACE2 binding while P681R in the furin cleavage site could increase the rate of S1-S2 cleavage, resulting in better transmissibility. The two RBD mutations, L452R and E484Q, indicated decreased binding to select monoclonal antibodies (mAbs) and may affect their neutralization potential. Further in vitro/in vivo studies would help confirm the phenotypic changes of the mutant strains. Overall, the study revealed that the newly emerged variants were responsible for the second wave of COVID-19 in Maharashtra. Lineage B.1.617.2 has been designated as a VOC delta and B.1.617.1 as a variant of interest kappa, and they are being widely reported in the rest of the country as well as globally. Continuous monitoring of these and emerging variants in India is essential.
Keywords: B.1.617.1; B.1.617.2; India; Maharashtra; SARS-CoV-2; evolution; modeling; second wave; whole genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

