Whole-genome sequencing of Schistosoma mansoni reveals extensive diversity with limited selection despite mass drug administration
- PMID: 34362894
- PMCID: PMC8346512
- DOI: 10.1038/s41467-021-24958-0
Whole-genome sequencing of Schistosoma mansoni reveals extensive diversity with limited selection despite mass drug administration
Abstract
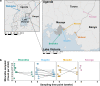
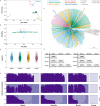
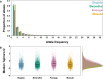
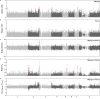
Control and elimination of the parasitic disease schistosomiasis relies on mass administration of praziquantel. Whilst these programmes reduce infection prevalence and intensity, their impact on parasite transmission and evolution is poorly understood. Here we examine the genomic impact of repeated mass drug administration on Schistosoma mansoni populations with documented reduced praziquantel efficacy. We sequenced whole-genomes of 198 S. mansoni larvae from 34 Ugandan children from regions with contrasting praziquantel exposure. Parasites infecting children from Lake Victoria, a transmission hotspot, form a diverse panmictic population. A single round of treatment did not reduce this diversity with no apparent population contraction caused by long-term praziquantel use. We find evidence of positive selection acting on members of gene families previously implicated in praziquantel action, but detect no high frequency functionally impactful variants. As efforts to eliminate schistosomiasis intensify, our study provides a foundation for genomic surveillance of this major human parasite.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- World Health Organization. Prevention and Control of Schistosomiasis and Soil-transmitted Helminthiasis: Report of a WHO Expert Committee (World Health Organization, 2002). - PubMed
-
- World Health Organization. Helminth Control in School-aged Children: A Guide for Managers of Control Programmes (World Health Organisation, 2006).
-
- World Health Organization. Schistosomiasis and soil-transmitted helminthiases: numbers of people treated in 2019. Wkly. Epidemiol. Rec. 2020;95:629–640.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

