Functional strain redundancy and persistent phage infection in Swiss hard cheese starter cultures
- PMID: 34363005
- PMCID: PMC8776748
- DOI: 10.1038/s41396-021-01071-0
Functional strain redundancy and persistent phage infection in Swiss hard cheese starter cultures
Abstract
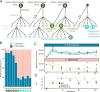
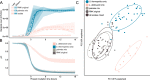
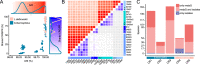
Undefined starter cultures are poorly characterized bacterial communities from environmental origin used in cheese making. They are phenotypically stable and have evolved through domestication by repeated propagation in closed and highly controlled environments over centuries. This makes them interesting for understanding eco-evolutionary dynamics governing microbial communities. While cheese starter cultures are known to be dominated by a few bacterial species, little is known about the composition, functional relevance, and temporal dynamics of strain-level diversity. Here, we applied shotgun metagenomics to an important Swiss cheese starter culture and analyzed historical and experimental samples reflecting 82 years of starter culture propagation. We found that the bacterial community is highly stable and dominated by only a few coexisting strains of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. lactis. Genome sequencing, metabolomics analysis, and co-culturing experiments of 43 isolates show that these strains are functionally redundant, but differ tremendously in their phage resistance potential. Moreover, we identified two highly abundant Streptococcus phages that seem to stably coexist in the community without any negative impact on bacterial growth or strain persistence, and despite the presence of a large and diverse repertoire of matching CRISPR spacers. Our findings show that functionally equivalent strains can coexist in domesticated microbial communities and highlight an important role of bacteria-phage interactions that are different from kill-the-winner dynamics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Cogan TM, Hill C. Cheese: chemistry, physics and microbiology. Boston: Springer; 1993.
-
- Powell IB, Broome MC, Limsowtin GKY. Reference Module in Food Science. London: Elsevier; 2016.
-
- Powell IB, Broome MC, Limsowtin GKY. Encyclopedia of Dairy Sciences. Cambridge: Academic Press; 2011.

