Metabolomics reveals biomarkers in human urine and plasma to predict cytochrome P450 2D6 (CYP2D6) activity
- PMID: 34363609
- PMCID: PMC9290485
- DOI: 10.1111/bph.15651
Metabolomics reveals biomarkers in human urine and plasma to predict cytochrome P450 2D6 (CYP2D6) activity
Abstract
Background and purpose: Individualized assessment of cytochrome P450 2D6 (CYP2D6) activity is usually performed through phenotyping following administration of a probe drug to measure the enzyme's activity. To avoid any iatrogenic harm (allergic drug reaction, dosing error) related to the probe drug, the development of non-burdensome tools for real-time phenotyping of CYP2D6 could significantly contribute to precision medicine. This study focuses on the identification of markers of the CYP2D6 enzyme in human biofluids using an LC-high-resolution mass spectrometry-based metabolomic approach.
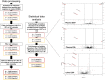
Experimental approach: Plasma and urine samples from healthy volunteers were analysed before and after intake of a daily dose of paroxetine 20 mg over 7 days. CYP2D6 genotyping and phenotyping, using single oral dose of dextromethorphan 5 mg, were also performed in all participants.
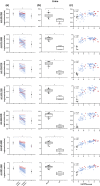
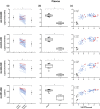
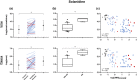
Key results: We report four metabolites of solanidine and two unknown compounds as possible novel CYP2D6 markers. Mean relative intensities of these features were significantly reduced during the inhibition session compared with the control session (n = 37). Semi-quantitative analysis showed that the largest decrease (-85%) was observed for the ion m/z 432.3108 normalized to solanidine (m/z 398.3417). Mean relative intensities of these ions were significantly higher in the CYP2D6 normal-ultrarapid metabolizer group (n = 37) compared with the poor metabolizer group (n = 6). Solanidine intensity was more than 15 times higher in CYP2D6-deficient individuals compared with other volunteers.
Conclusion and implications: The applied untargeted metabolomic strategy identified potential novel markers capable of semi-quantitatively predicting CYP2D6 activity, a promising discovery for personalized medicine.
Keywords: CYP2D6; CYP450; biomarker; metabolomics; phenotyping.
© 2021 The Authors. British Journal of Pharmacology published by John Wiley & Sons Ltd on behalf of British Pharmacological Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Sharman, J. L. , Southan, C. , Davies, J. A. , & CGTP Collaborators (2019). The Concise Guide to PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 - DOI - PMC - PubMed
-
- Beger, R. D. , Dunn, W. , Schmidt, M. A. , Gross, S. S. , Kirwan, J. A. , Cascante, M. , Brennan, L. , Wishart, D. S. , Oresic, M. , Hankemeier, T. , & Broadhurst, D. I. (2016). Metabolomics enables precision medicine: ‘A white paper, community perspective’. Metabolomics, 12, 149. 10.1007/s11306-016-1094-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

