Why are there so few (or so many) circulating coronaviruses?
- PMID: 34366247
- PMCID: PMC8272969
- DOI: 10.1016/j.it.2021.07.001
Why are there so few (or so many) circulating coronaviruses?
Abstract
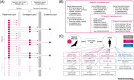
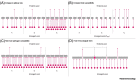
Despite vast diversity in non-human hosts and conspicuous recent spillover events, only a small number of coronaviruses have been observed to persist in human populations. This puzzling mismatch suggests substantial barriers to establishment. We detail hypotheses that might contribute to explain the low numbers of endemic coronaviruses, despite their considerable evolutionary and emergence potential. We assess possible explanations ranging from issues of ascertainment, historically lower opportunities for spillover, aspects of human demographic changes, and features of pathogen biology and pre-existing adaptive immunity to related viruses. We describe how successful emergent viral species must triangulate transmission, virulence, and host immunity to maintain circulation. Characterizing the factors that might shape the limits of viral persistence can delineate promising research directions to better understand the combinations of pathogens and contexts that are most likely to lead to spillover.
Keywords: antigenic space; cross-reactivity; landscape of immunity; viral ecology.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests None are declared.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

