Rapid Detection of New Delhi Metallo-β-Lactamase Gene Using Recombinase-Aided Amplification Directly on Clinical Samples From Children
- PMID: 34367092
- PMCID: PMC8339468
- DOI: 10.3389/fmicb.2021.691289
Rapid Detection of New Delhi Metallo-β-Lactamase Gene Using Recombinase-Aided Amplification Directly on Clinical Samples From Children
Abstract
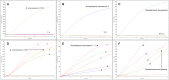
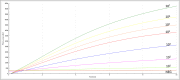
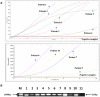
New Delhi metallo-β-lactamase, a metallo-β-lactamase carbapenemase type, mediates resistance to most β-lactam antibiotics including penicillins, cephalosporins, and carbapenems. Therefore, it is important to detect bla NDM genes in children's clinical samples as quickly as possible and analyze their characteristics. Here, a recombinase-aided amplification (RAA) assay, which operates in a single one-step reaction tube at 39°C in 5-15 min, was established to target bla NDM genes in children's clinical samples. The analytical sensitivity of the RAA assay was 20 copies, and the various bacterial types without bla NDM genes did not amplify. This method was used to detect bla NDM genes in 112 children's stool samples, 10 of which were tested positive by both RAA and standard PCR. To further investigate the characteristics of carbapenem-resistant bacteria carrying bla NDM in children, 15 carbapenem-resistant bacteria (Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Citrobacter freundii, Klebsiella oxytoca, Acinetobacter junii, and Proteus mirabilis) were isolated from the 10 samples. Notably, more than one bacterial type was isolated from three samples. Most of these isolates were resistant to cephalosporins, cefoperazone-sulbactam, piperacillin-tazobactam, ticarcillin-clavulanic acid, aztreonam, co-trimoxazole, and carbapenems. bla NDM - 1 and bla NDM - 5 were the two main types in these samples. These data show that the RAA assay has potential to be a sensitive and rapid bla NDM gene screening test for clinical samples. The common existence of bla NDM and multi-drug resistance genes presents major challenges for pediatric treatment.
Keywords: blaNDM; carbapenemase; character; pediatrics; recombinase-aided amplification.
Copyright © 2021 Feng, Xue, Feng, Yan, Cui, Gan, Zhang, Zhao, Xu, Li, Liu, Du, Zhang, Yao, Tai, Ma, Zhang, Qu, Wei and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Development of TaqMan real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii.Indian J Med Microbiol. 2011 Jul-Sep;29(3):249-53. doi: 10.4103/0255-0857.83907. Indian J Med Microbiol. 2011. PMID: 21860104
-
Detection of New Delhi Metallo-Beta-Lactamase-1 (NDM-1) in carbapenem- resistant Klebsiella pneumoniae isolated from a university hospital in Iran.Hippokratia. 2015 Jul-Sep;19(3):205-9. Hippokratia. 2015. PMID: 27418777 Free PMC article.
-
Resistotyping, phenotyping and genotyping of New Delhi metallo-β-lactamase (NDM) among Gram-negative bacilli from Iranian patients.J Med Microbiol. 2017 Apr;66(4):402-411. doi: 10.1099/jmm.0.000444. Epub 2017 Apr 21. J Med Microbiol. 2017. PMID: 28150578
-
Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health.BMC Microbiol. 2017 Apr 27;17(1):101. doi: 10.1186/s12866-017-1012-8. BMC Microbiol. 2017. PMID: 28449650 Free PMC article. Review.
-
NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings.Clin Microbiol Rev. 2019 Jan 30;32(2):e00115-18. doi: 10.1128/CMR.00115-18. Print 2019 Mar 20. Clin Microbiol Rev. 2019. PMID: 30700432 Free PMC article. Review.
Cited by
-
Rapid Detection of Multi-Resistance Strains Carrying mcr-1 Gene Using Recombinase-Aided Amplification Directly on Clinical Samples.Front Microbiol. 2022 Mar 31;13:852488. doi: 10.3389/fmicb.2022.852488. eCollection 2022. Front Microbiol. 2022. PMID: 35432248 Free PMC article.
-
Molecular epidemiology of carbapenemase encoding genes in A. baumannii-calcoaceticus complex infections in children: a systematic review.JAC Antimicrob Resist. 2024 Jul 12;6(4):dlae098. doi: 10.1093/jacamr/dlae098. eCollection 2024 Aug. JAC Antimicrob Resist. 2024. PMID: 39005591 Free PMC article.
-
Clinically relevant antibiotic resistance genes are linked to a limited set of taxa within gut microbiome worldwide.Nat Commun. 2023 Nov 14;14(1):7366. doi: 10.1038/s41467-023-42998-6. Nat Commun. 2023. PMID: 37963868 Free PMC article.
-
A Multiplex Recombinase-Aided qPCR Assay for Highly Sensitive and Rapid Detection of khe, blaKPC -2, and blaNDM -1 Genes in Klebsiella pneumoniae.J Clin Lab Anal. 2024 May;38(9):e25038. doi: 10.1002/jcla.25038. Epub 2024 Apr 8. J Clin Lab Anal. 2024. PMID: 38590133 Free PMC article.
-
Parallel Detection of the Unamplified Carbapenem Resistance Genes blaNDM-1 and blaOXA-1 Using a Plasmonic Nano-Biosensor with a Field-Portable DNA Extraction Method.Biosensors (Basel). 2025 Feb 14;15(2):112. doi: 10.3390/bios15020112. Biosensors (Basel). 2025. PMID: 39997014 Free PMC article.
References
-
- Bordin A., Trembizki E., Windsor M., Wee R., Tan L. Y., Buckley C., et al. (2019). Evaluation of the SpeeDx Carba (beta) multiplex real-time PCR assay for detection of NDM, KPC, OXA-48-like, IMP-4-like and VIM carbapenemase genes. BMC Infect. Dis. 19:571. 10.1186/s12879-019-4176-z - DOI - PMC - PubMed
-
- Dadashi M., Yaslianifard S., Hajikhani B., Kabir K., Owlia P., Goudarzi M., et al. (2019). Frequency distribution, genotypes and prevalent sequence types of New Delhi metallo-β-lactamase-producing Escherichia coli among clinical isolates around the world: a review. J. Glob. Antimicrob. Resist. 19 284–293. 10.1016/j.jgar.2019.06.008 - DOI - PubMed
-
- Fan G. H., Shen X. X., Li F., Li X. N., Bai X. D., Zhang R. Q., et al. (2019). Development of an Internally Controlled Reverse Transcription Recombinase-aided Amplification Assay for the Rapid and Visual Detection of West Nile Virus. Biomed. Environ. Sci. 32 926–929. 10.3967/bes2019.116 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

