Divergent Genomic Adaptations in the Microbiomes of Arctic Subzero Sea-Ice and Cryopeg Brines
- PMID: 34367102
- PMCID: PMC8339730
- DOI: 10.3389/fmicb.2021.701186
Divergent Genomic Adaptations in the Microbiomes of Arctic Subzero Sea-Ice and Cryopeg Brines
Abstract
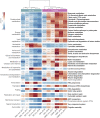
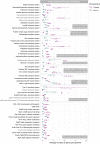
Subzero hypersaline brines are liquid microbial habitats within otherwise frozen environments, where concentrated dissolved salts prevent freezing. Such extreme conditions presumably require unique microbial adaptations, and possibly altered ecologies, but specific strategies remain largely unknown. Here we examined prokaryotic taxonomic and functional diversity in two seawater-derived subzero hypersaline brines: first-year sea ice, subject to seasonally fluctuating conditions; and ancient cryopeg, under relatively stable conditions geophysically isolated in permafrost. Overall, both taxonomic composition and functional potential were starkly different. Taxonomically, sea-ice brine communities (∼105 cells mL-1) had greater richness, more diversity and were dominated by bacterial genera, including Polaribacter, Paraglaciecola, Colwellia, and Glaciecola, whereas the more densely inhabited cryopeg brines (∼108 cells mL-1) lacked these genera and instead were dominated by Marinobacter. Functionally, however, sea ice encoded fewer accessory traits and lower average genomic copy numbers for shared traits, though DNA replication and repair were elevated; in contrast, microbes in cryopeg brines had greater genetic versatility with elevated abundances of accessory traits involved in sensing, responding to environmental cues, transport, mobile elements (transposases and plasmids), toxin-antitoxin systems, and type VI secretion systems. Together these genomic features suggest adaptations and capabilities of sea-ice communities manifesting at the community level through seasonal ecological succession, whereas the denser cryopeg communities appear adapted to intense bacterial competition, leaving fewer genera to dominate with brine-specific adaptations and social interactions that sacrifice some members for the benefit of others. Such cryopeg genomic traits provide insight into how long-term environmental stability may enable life to survive extreme conditions.
Keywords: cryopeg; cryosphere; hypersalinity; metagenomics; metatranscriptomics; microbial ecology; sea ice; subzero temperature.
Copyright © 2021 Rapp, Sullivan and Deming.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Andersen K. S., Kirkegaard R. H., Karst S. M., Albertsen M. (2018). ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 2018:299537. 10.1101/299537 - DOI
LinkOut - more resources
Full Text Sources

