CAR-NK Cells Effectively Target SARS-CoV-2-Spike-Expressing Cell Lines In Vitro
- PMID: 34367128
- PMCID: PMC8343231
- DOI: 10.3389/fimmu.2021.652223
CAR-NK Cells Effectively Target SARS-CoV-2-Spike-Expressing Cell Lines In Vitro
Abstract
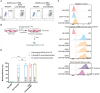
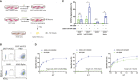
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is highly contagious and presents a significant public health issue. Current therapies used to treat coronavirus disease 2019 (COVID-19) include monoclonal antibody cocktail, convalescent plasma, antivirals, immunomodulators, and anticoagulants. The vaccines from Pfizer and Moderna have recently been authorized for emergency use, which are invaluable for the prevention of SARS-CoV-2 infection. However, their long-term side effects are not yet documented, and populations with immunocompromised conditions (e.g., organ-transplantation and immunodeficient patients) may not be able to mount an effective immune response. In addition, there are concerns that wide-scale immunity to SARS-CoV-2 may introduce immune pressure that could select for escape mutants to the existing vaccines and monoclonal antibody therapies. Emerging evidence has shown that chimeric antigen receptor (CAR)- natural killer (NK) immunotherapy has potent antitumor response in hematologic cancers with minimal adverse effects in recent studies, however, the potentials of CAR-NK cells in treating COVID-19 has not yet been fully exploited. Here, we improve upon a novel approach for the generation of CAR-NK cells for targeting SARS-CoV-2 and its various mutants. CAR-NK cells were generated using the scFv domain of S309 (henceforward, S309-CAR-NK), a SARS-CoV and SARS-CoV-2 neutralizing antibody (NAbs) that targets the highly conserved region of SARS-CoV-2 spike (S) glycoprotein and is therefore more likely to recognize different variants of SARS-CoV-2 isolates. S309-CAR-NK cells can specifically bind to pseudotyped SARS-CoV-2 virus and its D614G, N501Y, and E484K mutants. Furthermore, S309-CAR-NK cells can specifically kill target cells expressing SARS-CoV-2 S protein in vitro and show superior killing activity and cytokine production, compared to that of the recently reported CR3022-CAR-NK cells. Thus, these results pave the way for generating 'off-the-shelf' S309-CAR-NK cells for treatment in high-risk individuals as well as provide an alternative strategy for patients unresponsive to current vaccines.
Keywords: CAR (chimeric antigen receptor); COVID-19; E484K variant; N501Y variant; NK cells; SARS-CoV-2; off-the-shelf.
Copyright © 2021 Ma, Badeti, Chen, Kim, Choudhary, Honnen, Reichman, Calianese, Pinter, Jiang, Shi, Zhou, Xu, Li, Gause and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Update of
-
CAR-NK Cells Effectively Target the D614 and G614 SARS-CoV-2-infected Cells.bioRxiv [Preprint]. 2021 Jan 15:2021.01.14.426742. doi: 10.1101/2021.01.14.426742. bioRxiv. 2021. Update in: Front Immunol. 2021 Jul 23;12:652223. doi: 10.3389/fimmu.2021.652223. PMID: 33469580 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

