Immune2vec: Embedding B/T Cell Receptor Sequences in ℝ N Using Natural Language Processing
- PMID: 34367141
- PMCID: PMC8340020
- DOI: 10.3389/fimmu.2021.680687
Immune2vec: Embedding B/T Cell Receptor Sequences in ℝ N Using Natural Language Processing
Abstract
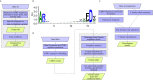
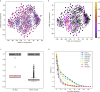
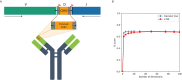
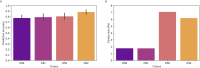
The adaptive branch of the immune system learns pathogenic patterns and remembers them for future encounters. It does so through dynamic and diverse repertoires of T- and B- cell receptors (TCR and BCRs, respectively). These huge immune repertoires in each individual present investigators with the challenge of extracting meaningful biological information from multi-dimensional data. The ability to embed these DNA and amino acid textual sequences in a vector-space is an important step towards developing effective analysis methods. Here we present Immune2vec, an adaptation of a natural language processing (NLP)-based embedding technique for BCR repertoire sequencing data. We validate Immune2vec on amino acid 3-gram sequences, continuing to longer BCR sequences, and finally to entire repertoires. Our work demonstrates Immune2vec to be a reliable low-dimensional representation that preserves relevant information of immune sequencing data, such as n-gram properties and IGHV gene family classification. Applying Immune2vec along with machine learning approaches to patient data exemplifies how distinct clinical conditions can be effectively stratified, indicating that the embedding space can be used for feature extraction and exploratory data analysis.
Keywords: BCR repertoire; NLP; biological sequence embedding; computational immunology; word2vec.
Copyright © 2021 Ostrovsky-Berman, Frankel, Polak and Yaari.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Murphy K. Janeway’s Immunobiology. 9. New York, NY: Garland Science; (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

