Single-Cell RNA Sequencing Reveals the Immunological Profiles of Renal Allograft Rejection in Mice
- PMID: 34367152
- PMCID: PMC8340022
- DOI: 10.3389/fimmu.2021.693608
Single-Cell RNA Sequencing Reveals the Immunological Profiles of Renal Allograft Rejection in Mice
Abstract
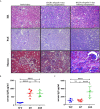
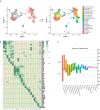
Allograft rejection is a common immunological feature in renal transplantation and is associated with reduced graft survival. A mouse renal allograft rejection model was induced and single-cell RNA sequencing (scRNA-seq) data of CD45+ leukocytes in kidney allografts on days 7 (D7) and 15 (D15) after operation was analyzed to reveal a full immunological profiling. We identified 20 immune cell types among 10,921 leukocytes. Macrophages and CD8+ T cells constituted the main populations on both timepoints. In the process from acute rejection (AR) towards chronic rejection (CR), the proportion of proliferating and naïve CD8+ T cells dropped significantly. Both B cells and neutrophils decreased by about 3 folds. On the contrary, the proportion of macrophages and dendritic cells (DCs) increased significantly, especially by about a 4.5-fold increase in Ly6cloMrc1+ macrophages and 2.6 folds increase in Ly6cloEar2+ macrophages. Moreover, myeloid cells harbored the richest ligand and receptor (LR) pairs with other cells, particularly for chemokine ligands such as Cxcl9, Cxcl10, Cxcl16 and Yars. However, macrophages with weak response to interferon gamma (IFNg) contributed to rejection chronicization. To conclude, reduction in CD8 T cells, B cells and neutrophils while increasing in Ly6cloMrc1+ macrophages and Ly6cloEar2+ macrophages, may contribute significantly to the progress from AR towards CR.
Keywords: ScRNA-seq; acute rejection; chronic rejection; immunological profiles; renal transplantation.
Copyright © 2021 Shen, Wang, Chen, Ma, Huang, Tang, Lan, Jiang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

