IRF7-Associated Immunophenotypes Have Dichotomous Responses to Virus/Allergen Coexposure and OM-85-Induced Reprogramming
- PMID: 34367159
- PMCID: PMC8339879
- DOI: 10.3389/fimmu.2021.699633
IRF7-Associated Immunophenotypes Have Dichotomous Responses to Virus/Allergen Coexposure and OM-85-Induced Reprogramming
Abstract
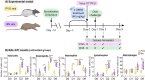
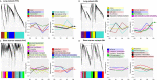
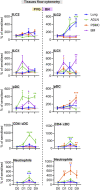
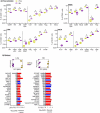
High risk for virus-induced asthma exacerbations in children is associated with an IRF7lo immunophenotype, but the underlying mechanisms are unclear. Here, we applied a Systems Biology approach to an animal model comprising rat strains manifesting high (BN) versus low susceptibility (PVG) to experimental asthma, induced by virus/allergen coexposure, to elucidate the mechanism(s)-of-action of the high-risk asthma immunophenotype. We also investigated potential risk mitigation via pretreatment with the immune training agent OM-85. Virus/allergen coexposure in low-risk PVG rats resulted in rapid and transient airways inflammation alongside IRF7 gene network formation. In contrast, responses in high-risk BN rats were characterized by severe airways eosinophilia and exaggerated proinflammatory responses that failed to resolve, and complete absence of IRF7 gene networks. OM-85 had more profound effects in high-risk BN rats, inducing immune-related gene expression changes in lung at baseline and reducing exaggerated airway inflammatory responses to virus/allergen coexposure. In low-risk PVG rats, OM-85 boosted IRF7 gene networks in the lung but did not alter baseline gene expression or cellular influx. Distinct IRF7-associated asthma risk immunophenotypes have dichotomous responses to virus/allergen coexposure and respond differentially to OM-85 pretreatment. Extrapolating to humans, our findings suggest that the beneficial effects OM-85 pretreatment may preferentially target those in high-risk subgroups.
Keywords: IRF7; allergen; asthma exacerbation; immunomodulation; rhinovirus; systems biology.
Copyright © 2021 de Jong, Lauzon-Joset, Leffler, Serralha, Larcombe, Christophersen, Holt, Strickland and Bosco.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Atopy-Dependent and Independent Immune Responses in the Heightened Severity of Atopics to Respiratory Viral Infections: Rat Model Studies.Front Immunol. 2018 Aug 13;9:1805. doi: 10.3389/fimmu.2018.01805. eCollection 2018. Front Immunol. 2018. PMID: 30150981 Free PMC article.
-
Methods for Experimental Allergen Immunotherapy: Subcutaneous and Sublingual Desensitization in Mouse Models of Allergic Asthma.Methods Mol Biol. 2021;2223:295-335. doi: 10.1007/978-1-0716-1001-5_20. Methods Mol Biol. 2021. PMID: 33226602
-
Effects of conventional tobacco smoke and nicotine-free cigarette smoke on airway inflammation, airway remodelling and lung function in a triple allergen model of severe asthma.Clin Exp Allergy. 2016 Jul;46(7):957-72. doi: 10.1111/cea.12665. Epub 2016 May 3. Clin Exp Allergy. 2016. PMID: 26502779
-
Virus/Allergen Interaction in Asthma Exacerbation.Ann Am Thorac Soc. 2015 Nov;12 Suppl 2(Suppl 2):S137-43. doi: 10.1513/AnnalsATS.201503-153AW. Ann Am Thorac Soc. 2015. PMID: 26595729 Free PMC article. Review.
-
Immune Responses in Rhinovirus-Induced Asthma Exacerbations.Curr Allergy Asthma Rep. 2016 Nov;16(11):78. doi: 10.1007/s11882-016-0661-2. Curr Allergy Asthma Rep. 2016. PMID: 27796793 Free PMC article. Review.
Cited by
-
LPS binding protein and activation signatures are upregulated during asthma exacerbations in children.Respir Res. 2023 Jul 12;24(1):184. doi: 10.1186/s12931-023-02478-3. Respir Res. 2023. PMID: 37438758 Free PMC article.
-
Emerging role for interferons in respiratory viral infections and childhood asthma.Front Immunol. 2023 Feb 21;14:1109001. doi: 10.3389/fimmu.2023.1109001. eCollection 2023. Front Immunol. 2023. PMID: 36895568 Free PMC article.
-
Oral Bacterial Lysate OM-85: Advances in Pharmacology and Therapeutics.Drug Des Devel Ther. 2024 Oct 1;18:4387-4399. doi: 10.2147/DDDT.S484897. eCollection 2024. Drug Des Devel Ther. 2024. PMID: 39372675 Free PMC article. Review.
-
Airway Administration of Bacterial Lysate OM-85 Protects Mice Against Respiratory Syncytial Virus Infection.Front Immunol. 2022 May 5;13:867022. doi: 10.3389/fimmu.2022.867022. eCollection 2022. Front Immunol. 2022. PMID: 35603159 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

