Ultra-High-Density QTL Marker Mapping for Seedling Photomorphogenesis Mediating Arabidopsis Establishment in Southern Patagonia
- PMID: 34367202
- PMCID: PMC8343176
- DOI: 10.3389/fpls.2021.677728
Ultra-High-Density QTL Marker Mapping for Seedling Photomorphogenesis Mediating Arabidopsis Establishment in Southern Patagonia
Abstract
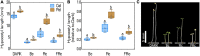
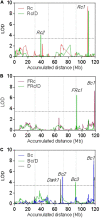
Arabidopsis thaliana shows a wide range of genetic and trait variation among wild accessions. Because of its unparalleled biological and genomic resources, Arabidopsis has a high potential for the identification of genes underlying ecologically important complex traits, thus providing new insights on genome evolution. Previous research suggested that distinct light responses were crucial for Arabidopsis establishment in a peculiar ecological niche of southern Patagonia. The aim of this study was to explore the genetic basis of contrasting light-associated physiological traits that may have mediated the rapid adaptation to this new environment. From a biparental cross between the photomorphogenic contrasting accessions Patagonia (Pat) and Columbia (Col-0), we generated a novel recombinant inbred line (RIL) population, which was entirely next-generation sequenced to achieve ultra-high-density saturating molecular markers resulting in supreme mapping sensitivity. We validated the quality of the RIL population by quantitative trait loci (QTL) mapping for seedling de-etiolation, finding seven QTLs for hypocotyl length in the dark and continuous blue light (Bc), continuous red light (Rc), and continuous far-red light (FRc). The most relevant QTLs, Rc1 and Bc1, were mapped close together to chromosome V; the former for Rc and Rc/dark, and the latter for Bc, FRc, and dark treatments. The additive effects of both QTLs were confirmed by independent heterogeneous inbred families (HIFs), and we explored TZP and ABA1 as potential candidate genes for Rc1 and Bc1QTLs, respectively. We conclude that the Pat × Col-0 RIL population is a valuable novel genetic resource to explore other adaptive traits in Arabidopsis.
Keywords: Arabidopsis; HIFs; Patagonia; QTL; RILs; seedling de-etiolation.
Copyright © 2021 Matsusaka, Filiault, Sanchez and Botto.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alonso-Blanco C., Peeters A. J. M., Koornneef M., Lister C., Dean C., Van Bosch N., et al. (1998). Development of an AFLP based linkage Map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14 259–271. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

