ExoS/ChvI Two-Component Signal-Transduction System Activated in the Absence of Bacterial Phosphatidylcholine
- PMID: 34367203
- PMCID: PMC8343143
- DOI: 10.3389/fpls.2021.678976
ExoS/ChvI Two-Component Signal-Transduction System Activated in the Absence of Bacterial Phosphatidylcholine
Abstract
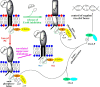
Sinorhizobium meliloti contains the negatively charged phosphatidylglycerol and cardiolipin as well as the zwitterionic phosphatidylethanolamine (PE) and phosphatidylcholine (PC) as major membrane phospholipids. In previous studies we had isolated S. meliloti mutants that lack PE or PC. Although mutants deficient in PE are able to form nitrogen-fixing nodules on alfalfa host plants, mutants lacking PC cannot sustain development of any nodules on host roots. Transcript profiles of mutants unable to form PE or PC are distinct; they differ from each other and they are different from the wild type profile. For example, a PC-deficient mutant of S. meliloti shows an increase of transcripts that encode enzymes required for succinoglycan biosynthesis and a decrease of transcripts required for flagellum formation. Indeed, a PC-deficient mutant is unable to swim and overproduces succinoglycan. Some suppressor mutants, that regain swimming and form normal levels of succinoglycan, are altered in the ExoS sensor. Our findings suggest that the lack of PC in the sinorhizobial membrane activates the ExoS/ChvI two-component regulatory system. ExoS/ChvI constitute a molecular switch in S. meliloti for changing from a free-living to a symbiotic life style. The periplasmic repressor protein ExoR controls ExoS/ChvI function and it is thought that proteolytic ExoR degradation would relieve repression of ExoS/ChvI thereby switching on this system. However, as ExoR levels are similar in wild type, PC-deficient mutant and suppressor mutants, we propose that lack of PC in the bacterial membrane provokes directly a conformational change of the ExoS sensor and thereby activation of the ExoS/ChvI two-component system.
Keywords: Sinorhizobium meliloti; membrane lipid; motility; phosphatidylethanolamine; succinoglycan; symbiosis.
Copyright © 2021 Geiger, Sohlenkamp, Vera-Cruz, Medeot, Martínez-Aguilar, Sahonero-Canavesi, Weidner, Pühler and López-Lara.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
 ), pcs-deficient mutant KDR568 (
), pcs-deficient mutant KDR568 ( ), PC-deficient mutant OG10017 (▲),PC-deficient mutant OG10017 harboring the empty plasmid pRK404 (○),PC-deficient mutant OG10017 complemented with pmtA-expressing plasmid pTB2042 (
), PC-deficient mutant OG10017 (▲),PC-deficient mutant OG10017 harboring the empty plasmid pRK404 (○),PC-deficient mutant OG10017 complemented with pmtA-expressing plasmid pTB2042 ( ), and PC-deficient mutant OG10017 complemented with pcs-expressing plasmid pTB2532 (
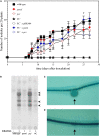
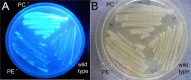
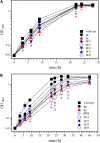
), and PC-deficient mutant OG10017 complemented with pcs-expressing plasmid pTB2532 ( ). The symbols of ▲ and ○ overlap. Values represent means of three independent experiments and standard deviation is shown. Statistical analysis was performed by a two-way ANOVA with Tukey’s multiple comparisons test as described in Materials and Methods. As an example, comparisons at 25 d after inoculation reveal that nodule numbers of all PC-deficient strains (PC–, PC– x pRK404) are significantly different (*p < 0.05) from PC-containing strains (wild type, pmtA–, pcs–, PC– x pmtA), except pcs– versus PC– or pcs– versus PC– x pRK404 (p < 0.0557). (B) PC-deficient S. meliloti mutant can form nodulation (Nod) factors. Thin-layer chromatographic analysis of Nod factor formation when S. meliloti wild type 1021 was not induced (lane 1) or induced with the flavonoid naringenin (lane 2), or when pmtA-deficient mutant KDR516 (lane 3), pcs-deficient mutant KDR568 (lane 4), or PC-deficient mutant OG10017 (lane 5) were induced with naringenin. All strains carried plasmid pMP280 for an increased production of Nod factors (Spaink et al., 1987). Arrowheads mark different Nod factors formed by S. meliloti. (C,D) PC-deficient S. meliloti mutant triggers initiation of nodule meristems on alfalfa roots. Arrows indicate a nodule induced by S. meliloti wild type (C), or a typical nodule meristem induced by the PC-deficient mutant OG10017 (D), respectively, 10 d after inoculation. For (C) and (D), roots were cleared with hypochlorite and stained with methylene blue as described by Truchet and collaborators (Truchet et al., 1989).
). The symbols of ▲ and ○ overlap. Values represent means of three independent experiments and standard deviation is shown. Statistical analysis was performed by a two-way ANOVA with Tukey’s multiple comparisons test as described in Materials and Methods. As an example, comparisons at 25 d after inoculation reveal that nodule numbers of all PC-deficient strains (PC–, PC– x pRK404) are significantly different (*p < 0.05) from PC-containing strains (wild type, pmtA–, pcs–, PC– x pmtA), except pcs– versus PC– or pcs– versus PC– x pRK404 (p < 0.0557). (B) PC-deficient S. meliloti mutant can form nodulation (Nod) factors. Thin-layer chromatographic analysis of Nod factor formation when S. meliloti wild type 1021 was not induced (lane 1) or induced with the flavonoid naringenin (lane 2), or when pmtA-deficient mutant KDR516 (lane 3), pcs-deficient mutant KDR568 (lane 4), or PC-deficient mutant OG10017 (lane 5) were induced with naringenin. All strains carried plasmid pMP280 for an increased production of Nod factors (Spaink et al., 1987). Arrowheads mark different Nod factors formed by S. meliloti. (C,D) PC-deficient S. meliloti mutant triggers initiation of nodule meristems on alfalfa roots. Arrows indicate a nodule induced by S. meliloti wild type (C), or a typical nodule meristem induced by the PC-deficient mutant OG10017 (D), respectively, 10 d after inoculation. For (C) and (D), roots were cleared with hypochlorite and stained with methylene blue as described by Truchet and collaborators (Truchet et al., 1989).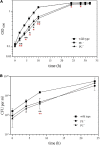


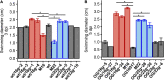
 ), and the correlated suppressor mutants M15 (
), and the correlated suppressor mutants M15 ( ), M16 (
), M16 ( ), M22 (
), M22 ( ), M33 (
), M33 ( ), M36 (
), M36 ( ) was recorded on LB/MC+ medium containing 20 mM Bis-Tris methane, pH 7.0 (A) or on LB/MC+ medium containing 20 mM Bis-Tris methane, pH 5.75 (B) by measuring the optical density of cultures at 600 nm (OD600). Values represent means of three independent experiments and standard deviation is shown. Statistical analysis was performed by a two-way ANOVA with Tukey’s multiple comparisons test as described in Materials and Methods. Comparisons of the PE-deficient (black asterisks) or the PC-deficient (red asterisks) mutant to wild type are indicated. Statistical significance is shown (*p < 0.05). Also indicated are statistically significant comparisons between the PC-deficient mutant and correlated suppressor mutants M16 (
) was recorded on LB/MC+ medium containing 20 mM Bis-Tris methane, pH 7.0 (A) or on LB/MC+ medium containing 20 mM Bis-Tris methane, pH 5.75 (B) by measuring the optical density of cultures at 600 nm (OD600). Values represent means of three independent experiments and standard deviation is shown. Statistical analysis was performed by a two-way ANOVA with Tukey’s multiple comparisons test as described in Materials and Methods. Comparisons of the PE-deficient (black asterisks) or the PC-deficient (red asterisks) mutant to wild type are indicated. Statistical significance is shown (*p < 0.05). Also indicated are statistically significant comparisons between the PC-deficient mutant and correlated suppressor mutants M16 ( ) and M36 (
) and M36 ( ).
).

Similar articles
-
ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti.Mol Microbiol. 2007 May;64(3):647-64. doi: 10.1111/j.1365-2958.2007.05680.x. Mol Microbiol. 2007. PMID: 17462014
-
Phosphatidylcholine-deficient suppressor mutant of Sinorhizobium meliloti, altered in fatty acid synthesis, partially recovers nodulation ability in symbiosis with alfalfa (Medicago sativa).Plant J. 2024 May;118(4):1136-1154. doi: 10.1111/tpj.16661. Epub 2024 Feb 11. Plant J. 2024. PMID: 38341846
-
Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis.Mol Microbiol. 2009 Dec;74(5):1223-37. doi: 10.1111/j.1365-2958.2009.06931.x. Epub 2009 Oct 19. Mol Microbiol. 2009. PMID: 19843226
-
Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions.Mol Plant Microbe Interact. 2003 Jul;16(7):567-79. doi: 10.1094/MPMI.2003.16.7.567. Mol Plant Microbe Interact. 2003. PMID: 12848422 Review.
-
Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance.Biochem Soc Trans. 1991 Aug;19(3):636-41. doi: 10.1042/bst0190636. Biochem Soc Trans. 1991. PMID: 1783190 Review.
Cited by
-
Three separate pathways in Rhizobium leguminosarum maintain phosphatidylcholine biosynthesis, which is required for symbiotic nitrogen fixation with clover.Appl Environ Microbiol. 2024 Sep 18;90(9):e0059024. doi: 10.1128/aem.00590-24. Epub 2024 Aug 9. Appl Environ Microbiol. 2024. PMID: 39120150 Free PMC article.
-
A sensor histidine kinase from a plant-endosymbiont bacterium restores the virulence of a mammalian intracellular pathogen.Microb Pathog. 2023 Dec;185:106442. doi: 10.1016/j.micpath.2023.106442. Epub 2023 Nov 8. Microb Pathog. 2023. PMID: 37944675 Free PMC article.
-
A protease and a lipoprotein jointly modulate the conserved ExoR-ExoS-ChvI signaling pathway critical in Sinorhizobium meliloti for symbiosis with legume hosts.PLoS Genet. 2023 Oct 23;19(10):e1010776. doi: 10.1371/journal.pgen.1010776. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37871041 Free PMC article.
-
Activation of ChvG-ChvI regulon by cell wall stress confers resistance to β-lactam antibiotics and initiates surface spreading in Agrobacterium tumefaciens.PLoS Genet. 2022 Dec 8;18(12):e1010274. doi: 10.1371/journal.pgen.1010274. eCollection 2022 Dec. PLoS Genet. 2022. PMID: 36480495 Free PMC article.
-
Recombinant and endogenous ways to produce methylated phospholipids in Escherichia coli.Appl Microbiol Biotechnol. 2021 Dec;105(23):8837-8851. doi: 10.1007/s00253-021-11654-8. Epub 2021 Oct 28. Appl Microbiol Biotechnol. 2021. PMID: 34709431 Free PMC article.
References
LinkOut - more resources
Full Text Sources

