The Added Value of Whole-Exome Sequencing for Anomalous Fetuses With Detailed Prenatal Ultrasound and Postnatal Phenotype
- PMID: 34367232
- PMCID: PMC8340955
- DOI: 10.3389/fgene.2021.627204
The Added Value of Whole-Exome Sequencing for Anomalous Fetuses With Detailed Prenatal Ultrasound and Postnatal Phenotype
Abstract
Objectives: The objective of the study was to explore the added value of whole-exome sequencing (WES) in abnormal fetuses with detailed prenatal ultrasound and postnatal phenotype with normal karyotype and chromosomal microarray analysis (CMA).
Methods: Parents of fetuses with structural abnormalities by prenatal ultrasound who consented to provide fetal samples were prospectively recruited from January 2017 to December 2019. With aneuploidies or cases with copy number variations (CNVs) excluded, WES was performed for cases with normal karyotype and CMA results. Detailed prenatal ultrasound and postnatal imaging or pathology features were recommended for further interpretation of genetic variants.
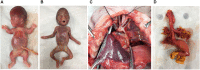
Results: WES was performed for 94 eligible fetuses, DNA samples of which were extracted from 53 parent-fetus trios and 41 proband-only fetal tissues. A diagnostic genetic variant was identified in 37 (39.4%) of 94 fetuses, and 34 (64.2%) were detected in 53 trios, which was significantly greater than 3 (7.3%) in 41 proband-only cases (p < 0.001). In 34 trios with diagnostic genetic variants, 23 (67.6%) were de novo and 11 (32.4%) were inherited with two homozygous and nine heterozygous variants. Fourteen (14.9%) of 94 fetuses had a variant of uncertain significance (VUS). Among 94 cases, six affected pregnancies continued and 88 terminated, and 57 of 88 terminated cases underwent postmortem examinations. With accurate phenotypes demonstrated by prenatal ultrasound and postnatal autopsies, the clinical phenotypes were correlated in 33 (89.2%) of 37 cases with specific genotypes, with the highest matching ratio in skeletal diseases (20/33, 60.6%).
Conclusion: WES has added value in the genetic diagnosis of abnormal fetuses with normal karyotypes and CMA, particularly in skeletal diseases. Using WES in various anomalous fetuses can broaden the understanding of prenatal phenotypes and genetic variants.
Keywords: anomalous fetuses; genetic variants; postnatal phenotype; prenatal ultrasound; whole-exome sequencing.
Copyright © 2021 He, Du, Xie, Zhang, Gu, Lei, Zheng and Chen.
Conflict of interest statement
DC was employed by the company Guangzhou Kingmed Diagnostics Group. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study.Lancet. 2019 Feb 23;393(10173):758-767. doi: 10.1016/S0140-6736(18)32042-7. Epub 2019 Jan 31. Lancet. 2019. PMID: 30712878
-
Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study.Lancet. 2019 Feb 23;393(10173):747-757. doi: 10.1016/S0140-6736(18)31940-8. Epub 2019 Jan 31. Lancet. 2019. PMID: 30712880 Free PMC article.
-
Whole exome sequencing as a diagnostic adjunct to clinical testing in fetuses with structural abnormalities.Ultrasound Obstet Gynecol. 2018 Apr;51(4):493-502. doi: 10.1002/uog.18915. Ultrasound Obstet Gynecol. 2018. PMID: 28976722
-
Exome Sequencing in Fetuses with Structural Malformations.J Clin Med. 2014 Jul 8;3(3):747-62. doi: 10.3390/jcm3030747. J Clin Med. 2014. PMID: 26237476 Free PMC article. Review.
-
Promises, pitfalls and practicalities of prenatal whole exome sequencing.Prenat Diagn. 2018 Jan;38(1):10-19. doi: 10.1002/pd.5102. Epub 2017 Jul 25. Prenat Diagn. 2018. PMID: 28654730 Free PMC article. Review.
Cited by
-
The challenges in the interpretation of genetic variants detected by genomics techniques in patients with congenital anomalies.J Clin Lab Anal. 2023 Sep;37(17-18):e24967. doi: 10.1002/jcla.24967. Epub 2023 Oct 12. J Clin Lab Anal. 2023. PMID: 37823350 Free PMC article.
-
Not putting the cart before the horse: the complex social and ethical terrain of prenatal exome sequencing.Eur J Hum Genet. 2023 Feb;31(2):134-135. doi: 10.1038/s41431-022-01225-4. Epub 2022 Nov 7. Eur J Hum Genet. 2023. PMID: 36336716 Free PMC article. No abstract available.
-
Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: A systematic review and meta-analysis.Prenat Diagn. 2022 May;42(6):662-685. doi: 10.1002/pd.6115. Epub 2022 May 7. Prenat Diagn. 2022. PMID: 35170059 Free PMC article.
-
Exome sequencing in every pregnancy? Results of trio exome sequencing in structurally normal fetuses.Prenat Diagn. 2025 Mar;45(3):276-286. doi: 10.1002/pd.6585. Epub 2024 May 12. Prenat Diagn. 2025. PMID: 38735835 Free PMC article.
-
The new frontier: a case for whole exome sequencing with multiple fetal anomalies.Case Rep Perinat Med. 2023 May 4;12(1):20220032. doi: 10.1515/crpm-2022-0032. eCollection 2023 Jan. Case Rep Perinat Med. 2023. PMID: 40041274 Free PMC article.
References
-
- Chandler N., Best S., Hayward J., Faravelli F., Mansour S., Kivuva E., et al. (2018). Rapid prenatal diagnosis using targeted exome sequencing: a cohort study to assess feasibility and potential impact on prenatal counseling and pregnancy management. Genet. Med. 20 1430–1437. 10.1038/gim.2018.30 - DOI - PubMed
LinkOut - more resources
Full Text Sources

