A systems-based framework to computationally describe putative transcription factors and signaling pathways regulating glycan biosynthesis
- PMID: 34367349
- PMCID: PMC8313979
- DOI: 10.3762/bjoc.17.119
A systems-based framework to computationally describe putative transcription factors and signaling pathways regulating glycan biosynthesis
Abstract
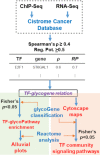
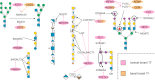
Glycosylation is a common posttranslational modification, and glycan biosynthesis is regulated by a set of glycogenes. The role of transcription factors (TFs) in regulating the glycogenes and related glycosylation pathways is largely unknown. In this work, we performed data mining of TF-glycogene relationships from the Cistrome Cancer database (DB), which integrates chromatin immunoprecipitation sequencing (ChIP-Seq) and RNA-Seq data to constitute regulatory relationships. In total, we observed 22,654 potentially significant TF-glycogene relationships, which include interactions involving 526 unique TFs and 341 glycogenes that span 29 the Cancer Genome Atlas (TCGA) cancer types. Here, TF-glycogene interactions appeared in clusters or so-called communities, suggesting that changes in single TF expression during both health and disease may affect multiple carbohydrate structures. Upon applying the Fisher's exact test along with glycogene pathway classification, we identified TFs that may specifically regulate the biosynthesis of individual glycan types. Integration with Reactome DB knowledge provided an avenue to relate cell-signaling pathways to TFs and cellular glycosylation state. Whereas analysis results are presented for all 29 cancer types, specific focus is placed on human luminal and basal breast cancer disease progression. Overall, the article presents a computational approach to describe TF-glycogene relationships, the starting point for experimental system-wide validation.
Keywords: ChIP-Seq; TCGA transcription factor; glycoinformatics; glycosylation.
Copyright © 2021, Groth et al.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
