Numerical simulation of intracellular drug delivery via rapid squeezing
- PMID: 34367404
- PMCID: PMC8331209
- DOI: 10.1063/5.0059165
Numerical simulation of intracellular drug delivery via rapid squeezing
Abstract
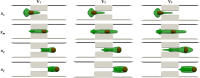
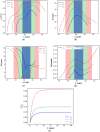
Intracellular drug delivery by rapid squeezing is one of the most recent and simple cell membrane disruption-mediated drug encapsulation approaches. In this method, cell membranes are perforated in a microfluidic setup due to rapid cell deformation during squeezing through constricted channels. While squeezing-based drug loading has been successful in loading drug molecules into various cell types, such as immune cells, cancer cells, and other primary cells, there is so far no comprehensive understanding of the pore opening mechanism on the cell membrane and the systematic analysis on how different channel geometries and squeezing speed influence drug loading. This article aims to develop a three-dimensional computational model to study the intracellular delivery for compound cells squeezing through microfluidic channels. The Lattice Boltzmann method, as the flow solver, integrated with a spring-connected network via frictional coupling, is employed to capture compound capsule dynamics over fast squeezing. The pore size is proportional to the local areal strain of triangular patches on the compound cell through mathematical correlations derived from molecular dynamics and coarse-grained molecular dynamics simulations. We quantify the drug concentration inside the cell cytoplasm by introducing a new mathematical model for passive diffusion after squeezing. Compared to the existing models, the proposed model does not have any empirical parameters that depend on operating conditions and device geometry. Since the compound cell model is new, it is validated by simulating a nucleated cell under a simple shear flow at different capillary numbers and comparing the results with other numerical models reported in literature. The cell deformation during squeezing is also compared with the pattern found from our compound cell squeezing experiment. Afterward, compound cell squeezing is modeled for different cell squeezing velocities, constriction lengths, and constriction widths. We reported the instantaneous cell center velocity, variations of axial and vertical cell dimensions, cell porosity, and normalized drug concentration to shed light on the underlying physics in fast squeezing-based drug delivery. Consistent with experimental findings in the literature, the numerical results confirm that constriction width reduction, constriction length enlargement, and average cell velocity promote intracellular drug delivery. The results show that the existence of the nucleus increases cell porosity and loaded drug concentration after squeezing. Given geometrical parameters and cell average velocity, the maximum porosity is achieved at three different locations: constriction entrance, constriction middle part, and outside the constriction. Our numerical results provide reasonable justifications for experimental findings on the influences of constriction geometry and cell velocity on the performance of cell-squeezing delivery. We expect this model can help design and optimize squeezing-based cargo delivery.
© 2021 Author(s).
Figures










References
-
- Kitao T. and Hattori K., “Erythrocyte entrapment of daunomycin by amphotericin B without hemolysis,” Cancer Res. 40(4), 1351–1353 (1980). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
