Larger cells have relatively smaller nuclei across the Tree of Life
- PMID: 34367657
- PMCID: PMC8327945
- DOI: 10.1002/evl3.243
Larger cells have relatively smaller nuclei across the Tree of Life
Abstract
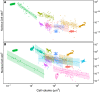
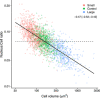
Larger cells have larger nuclei, but the precise relationship between cell size and nucleus size remains unclear, and the evolutionary forces that shape this relationship are debated. We compiled data for almost 900 species - from yeast to mammals - at three scales of biological organisation: among-species, within-species, and among-lineages of a species that was artificially selected for cell size. At all scales, we showed that the ratio of nucleus size to cell size (the 'N: C' ratio) decreased systematically in larger cells. Size evolution appears more constrained in nuclei than cells: cell size spans across six orders of magnitude, whereas nucleus size varies by only three. The next important challenge is to determine the drivers of this apparently ubiquitous relationship in N:C ratios across such a diverse array of organisms.
Keywords: Artificial selection; C‐value enigma; genome size evolution; karyoplasmic ratio; limiting pool hypothesis; nucleoskeletal theory; nucleotypic theory; optimal DNA theory; selfish DNA hypothesis.
© 2021 The Authors. Evolution Letters published by Wiley Periodicals LLC on behalf of Society for the Study of Evolution (SSE) and European Society for Evolutionary Biology (ESEB).
Figures

References
-
- Bates, D. , Mächler M., Bolker B., and Walker S. 2015. Fitting Linear Mixed‐Effects Models Usinglme4. Journal of Statistical Software 67: 1–48.
-
- Brett, M. T. 2004. When is a correlation between non‐independent variables “spurious”? Oikos 105: 647–656.
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

