The sRNA Regulated Protein DdbA Is Involved in Development and Maintenance of the Chlamydia trachomatis EB Cell Form
- PMID: 34368013
- PMCID: PMC8343073
- DOI: 10.3389/fcimb.2021.692224
The sRNA Regulated Protein DdbA Is Involved in Development and Maintenance of the Chlamydia trachomatis EB Cell Form
Abstract
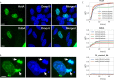
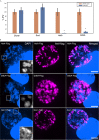
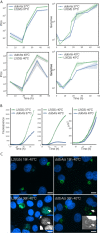
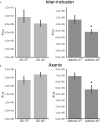
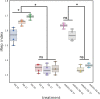
The chlamydial small non coding RNA, IhtA, regulates the expression of both HctA and DdbA, the uncharacterized product of the C. trachomatis L2 CTL0322 gene. HctA is a small, highly basic, DNA binding protein that is expressed late in development and mediates the condensation of the genome during RB to EB differentiation. DdbA is conserved throughout the chlamydial lineage, and is predicted to express a small, basic, cytoplasmic protein. As it is common for sRNAs to regulate multiple mRNAs within the same physiological pathway, we hypothesize that DdbA, like HctA, is involved in RB to EB differentiation. Here, we show that DdbA is a DNA binding protein, however unlike HctA, DdbA does not contribute to genome condensation but instead likely has nuclease activity. Using a DdbA temperature sensitive mutant, we show that DdbAts creates inclusions indistinguishable from WT L2 in size and that early RB replication is likewise similar at the nonpermissive temperature. However, the number of DdbAts infectious progeny is dramatically lower than WT L2 overall, although production of EBs is initiated at a similar time. The expression of a late gene reporter construct followed live at 40°C indicates that late gene expression is severely compromised in the DdbAts strain. Viability assays, both in host cells and in axenic media indicate that the DdbAts strain is defective in the maintenance of EB infectivity. Additionally, using Whole Genome Sequencing we demonstrate that chromosome condensation is temporally separated from DNA replication during the RB to EB transition. Although DdbA does not appear to be directly involved in this process, our data suggest it is a DNA binding protein that is important in the production and maintenance of infectivity of the EB, perhaps by contributing to the remodeling of the EB chromosome.
Keywords: Chlamydia; bacterial cell development; bacterial replication; elementary bodies; reticulate body.
Copyright © 2021 Grieshaber, Runac, Turner, Dean, Appa, Omsland and Grieshaber.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Translation inhibition of the developmental cycle protein HctA by the small RNA IhtA is conserved across Chlamydia.PLoS One. 2012;7(10):e47439. doi: 10.1371/journal.pone.0047439. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071807 Free PMC article.
-
Impact of Active Metabolism on Chlamydia trachomatis Elementary Body Transcript Profile and Infectivity.J Bacteriol. 2018 Jun 25;200(14):e00065-18. doi: 10.1128/JB.00065-18. Print 2018 Jul 15. J Bacteriol. 2018. PMID: 29735758 Free PMC article.
-
Identification of the base-pairing requirements for repression of hctA translation by the small RNA IhtA leads to the discovery of a new mRNA target in Chlamydia trachomatis.PLoS One. 2015 Mar 10;10(3):e0116593. doi: 10.1371/journal.pone.0116593. eCollection 2015. PLoS One. 2015. PMID: 25756658 Free PMC article.
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
One Face of Chlamydia trachomatis: The Infectious Elementary Body.Curr Top Microbiol Immunol. 2018;412:35-58. doi: 10.1007/82_2016_12. Curr Top Microbiol Immunol. 2018. PMID: 27197644 Review.
Cited by
-
The T3SS structural and effector genes of Chlamydia trachomatis are expressed in distinct phenotypic cell forms.Front Cell Infect Microbiol. 2025 May 8;15:1579247. doi: 10.3389/fcimb.2025.1579247. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40406518 Free PMC article.
-
The chlamydial transcriptional regulator Euo is a key switch in cell form developmental progression but is not involved in the committed step to the formation of the infectious form.mSphere. 2024 Sep 25;9(9):e0043724. doi: 10.1128/msphere.00437-24. Epub 2024 Aug 14. mSphere. 2024. PMID: 39140730 Free PMC article.
-
Recent advances in genetic systems in obligate intracellular human-pathogenic bacteria.Front Cell Infect Microbiol. 2023 Jun 19;13:1202245. doi: 10.3389/fcimb.2023.1202245. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37404720 Free PMC article. Review.
-
Translational gene expression control in Chlamydia trachomatis.PLoS One. 2022 Jan 27;17(1):e0257259. doi: 10.1371/journal.pone.0257259. eCollection 2022. PLoS One. 2022. PMID: 35085261 Free PMC article.
-
Pathogenicity and virulence of Chlamydia trachomatis: Insights into host interactions, immune evasion, and intracellular survival.Virulence. 2025 Dec;16(1):2503423. doi: 10.1080/21505594.2025.2503423. Epub 2025 May 15. Virulence. 2025. PMID: 40353442 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

