Coronary Artery Disease Genetics Enlightened by Genome-Wide Association Studies
- PMID: 34368511
- PMCID: PMC8326228
- DOI: 10.1016/j.jacbts.2021.04.001
Coronary Artery Disease Genetics Enlightened by Genome-Wide Association Studies
Abstract
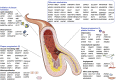
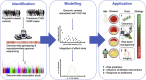
Many cardiovascular diseases are facilitated by strong inheritance. For example, large-scale genetic studies identified hundreds of genomic loci that affect the risk of coronary artery disease. At each of these loci, common variants are associated with disease risk with robust statistical evidence but individually small effect sizes. Only a minority of candidate genes found at these loci are involved in the pathophysiology of traditional risk factors, but experimental research is making progress in identifying novel, and, in part, unexpected mechanisms. Targets identified by genome-wide association studies have already led to the development of novel treatments, specifically in lipid metabolism. This review summarizes recent genetic and experimental findings in this field. In addition, the development and possible clinical usefulness of polygenic risk scores in risk prediction and individualization of treatment, particularly in lipid metabolism, are discussed.
Keywords: CAD, coronary artery disease; CXCL1, chemokine (C-X-C motif) ligand 1; GWAS, genome-wide association study; LDLR, low-density lipoprotein receptor; LPL, lipoprotein lipase; MI, myocardial infarction; PCSK9, proprotein convertase subtilisin/kexin type 9; cardiovascular diseases; coronary artery disease; genome-wide association studies; lncRNA, long non-coding RNA; polygenic risk scores; precision medicine.
© 2021 The Authors.
Conflict of interest statement
Dr. Kessler was supported by the Corona Foundation as part of the Junior Research Group Translational Cardiovascular Genomics (S199/10070/2017) and the German Research Foundation as part of the collaborative research center SFB 1123 (B02) Dr. Schunkert was supported by German Research Foundation as part of the collaborative research centers SFB 1123 TRR 267 (B06) and also supported by additional grants received from the German Federal Ministry of Education and Research within the framework of ERA-NET on Cardiovascular Disease (ERA-CVD: grant JTC2017_21-040) and within the scheme of target validation (BlockCAD: 6GW0198K). Further support was received from the British Heart Foundation/DZHK collaborative project “Genetic discovery-based targeting of the vascular interface in atherosclerosis.” Dr. Schunkert has received personal fees from MSD Sharp & Dohme, Amgen, Bayer Vital GmbH, Boehringer Ingelheim, Daiichi-Sankyo, Novartis, Servier, Brahms, Bristol-Myers-Squibb, Medtronic, Sanofi Aventis, Synlab, Pfizer, and Vifor T as well as grants and personal fees from Astra-Zeneca outside the submitted work. Drs. Schunkert and Kessler are named inventors on a patent application for prevention of restenosis after angioplasty and stent implantation outside the submitted work.
Figures


References
-
- Benjamin E.J., Muntner P., Alonso A. Heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation. 2019;139:e56–e528. - PubMed
-
- Schunkert H., Scheidt von M., Kessler T., Stiller B., Zeng L., Vilne B. Genetics of coronary artery disease in the light of genome-wide association studies. Clin Res Cardiol. 2018;120:963–968. - PubMed
-
- Yusuf S., Hawken S., Ounpuu S. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–952. - PubMed
-
- Marenberg M.E., Risch N., Berkman L.F., Floderus B., de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. - PubMed
-
- Mayer B., Erdmann J., Schunkert H. Genetics and heritability of coronary artery disease and myocardial infarction. Clin Res Cardiol. 2007;96:1–7. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

