PAK4 inhibition improves PD-1 blockade immunotherapy
- PMID: 34368780
- PMCID: PMC8340852
- DOI: 10.1038/s43018-019-0003-0
PAK4 inhibition improves PD-1 blockade immunotherapy
Erratum in
-
Publisher Correction: PAK4 inhibition improves PD-1 blockade immunotherapy.Nat Cancer. 2020 Feb;1(2):264. doi: 10.1038/s43018-020-0025-7. Nat Cancer. 2020. PMID: 35122015 No abstract available.
Abstract
Lack of tumor infiltration by immune cells is the main mechanism of primary resistance to programmed cell death protein 1 (PD-1) blockade therapies for cancer. It has been postulated that cancer cell-intrinsic mechanisms may actively exclude T cells from tumors, suggesting that the finding of actionable molecules that could be inhibited to increase T cell infiltration may synergize with checkpoint inhibitor immunotherapy. Here, we show that p21-activated kinase 4 (PAK4) is enriched in non-responding tumor biopsies with low T cell and dendritic cell infiltration. In mouse models, genetic deletion of PAK4 increased T cell infiltration and reversed resistance to PD-1 blockade in a CD8 T cell-dependent manner. Furthermore, combination of anti-PD-1 with the PAK4 inhibitor KPT-9274 improved anti-tumor response compared with anti-PD-1 alone. Therefore, high PAK4 expression is correlated with low T cell and dendritic cell infiltration and a lack of response to PD-1 blockade, which could be reversed with PAK4 inhibition.
Conflict of interest statement
Competing interests G.A.-R. has received honoraria for consulting with Arcus Biosciences. W.S. and E.B. were employees of Karyopharm Therapeutics when this study was conducted. A.R. has received honoraria for consulting with Amgen, Bristol-Myers Squibb, Chugai, Genentech, Merck, Novartis, Roche and Sanofi, is or has been a member of the scientific advisory board, and holds stock in Advaxis, Arcus Biosciences, Bioncotech Therapeutics, Compugen, CytomX, Five Prime, FLX Bio, ImaginAb, IsoPlexis, Gilead Kite, Lutris Pharma, Merus, PACT Pharma, Rgenix and Tango Therapeutics. G.A.-R., D.Y.T., C.S.G. and A.R. are inventors in a patent application covering the use of PAK4 inhibitors for cancer immunotherapy.
Figures









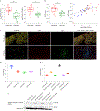


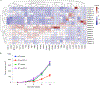

Comment in
-
PAK4 as a cancer immune-evasion target.Nat Cancer. 2020 Jan;1(1):18-19. doi: 10.1038/s43018-019-0012-z. Nat Cancer. 2020. PMID: 35121838 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

