An optimized, amplicon-based approach for sequencing of SARS-CoV-2 from patient samples using COVIDSeq assay on Illumina MiSeq sequencing platforms
- PMID: 34368787
- PMCID: PMC8326003
- DOI: 10.1016/j.xpro.2021.100755
An optimized, amplicon-based approach for sequencing of SARS-CoV-2 from patient samples using COVIDSeq assay on Illumina MiSeq sequencing platforms
Abstract
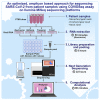
Sequencing of SARS-CoV-2 genomes is crucial for understanding the genetic epidemiology of the COVID-19 pandemic. It is also critical for understanding the evolution of the virus and also for the rapid development of diagnostic tools. The present protocol is a modification of the Illumina COVIDSeq test. We describe an amplicon-based next-generation sequencing approach with short turnaround time, adapted for bench-top sequencers like MiSeq, iSeq, and MiniSeq. For complete details on the use and execution of this protocol, please refer to Bhoyar et al. (2021).
Keywords: Bioinformatics; Clinical Protocol; Genomics; Immunology; Microbiology; Sequence analysis; Sequencing.
© 2021.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Poojary M., Shantaraman A., Jolly B., Scaria V. Computational protocol for assembly and analysis of SARS-nCoV-2 genomes. Res. Rep. 2020;4:e1–e14doi. doi: 10.9777/rr.2020.10001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

