Abnormal migration behavior linked to Rac1 signaling contributes to primordial germ cell exhaustion in Fanconi anemia pathway-deficient Fancg-/- embryos
- PMID: 34368842
- PMCID: PMC8682768
- DOI: 10.1093/hmg/ddab222
Abnormal migration behavior linked to Rac1 signaling contributes to primordial germ cell exhaustion in Fanconi anemia pathway-deficient Fancg-/- embryos
Abstract
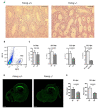
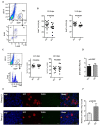
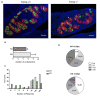
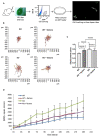
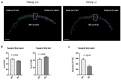
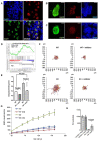
Fanconi anemia (FA) is a rare human genetic disorder characterized by bone marrow failure, predisposition to cancer and developmental defects including hypogonadism. Reproductive defects leading to germ cell aplasia are the most consistent phenotypes seen in FA mouse models. We examined the role of the nuclear FA core complex gene Fancg in the development of primordial germ cells (PGCs), the embryonic precursors of adult gametes, during fetal development. PGC maintenance was severely impaired in Fancg-/- embryos. We observed a defect in the number of PGCs starting at E9.5 and a strong attrition at E11.5 and E13.5. Remarkably, we observed a mosaic pattern reflecting a portion of testicular cords devoid of PGCs in E13.5 fetal gonads. Our in vitro and in vivo data highlight a potential role of Fancg in the proliferation and in the intrinsic cell motility abilities of PGCs. The random migratory process is abnormally activated in Fancg-/- PGCs, altering the migration of cells. Increased cell death and PGC attrition observed in E11.5 Fancg-/- embryos are features consistent with delayed migration of PGCs along the migratory pathway to the genital ridges. Moreover, we show that an inhibitor of RAC1 mitigates the abnormal migratory pattern observed in Fancg-/- PGCs.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- Ceccaldi, R., Sarangi, P. and D’Andrea, A.D. (2016) The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol., 17, 337–349. - PubMed
-
- Blom, E., van de Vrugt, H.J., de Vries, Y., de Winter, J.P., Arwert, F. and Joenje, H. (2004) Multiple TPR motifs characterize the Fanconi anemia FANCG protein. DNA Repair (Amst), 3, 77–84. - PubMed
-
- Blanpain, C., Mohrin, M., Sotiropoulou, P.A. and Passegue, E. (2011) DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell, 8, 16–29. - PubMed
-
- Haneline, L.S., Gobbett, T.A., Ramani, R., Carreau, M., Buchwald, M., Yoder, M.C. and Clapp, D.W. (1999) Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood, 94, 1–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

