Disentangling tumorigenesis-associated DNA methylation changes in colorectal tissues from those associated with ageing
- PMID: 34369258
- PMCID: PMC9235894
- DOI: 10.1080/15592294.2021.1952375
Disentangling tumorigenesis-associated DNA methylation changes in colorectal tissues from those associated with ageing
Abstract
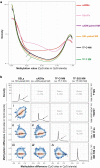
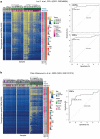
Physiological ageing and tumorigenesis are both associated with epigenomic alterations in human tissue cells, the most extensively investigated of which entails de novo cytosine methylation (i.e., hypermethylation) within the CpG dinucleotides of CpG islands. Genomic regions that become hypermethylated during tumorigenesis are generally believed to overlap regions that acquire methylation in normal tissues as an effect of ageing. To define the extension of this overlap, we analysed the DNA methylomes of 48 large-bowel tissue samples taken from women of different ages during screening colonoscopy: 18 paired samples of normal and lesional tissues from donors harbouring a precancerous lesion and 12 samples of normal mucosa from tumour-free donors. Each sample was subjected to targeted, genome-wide bisulphite sequencing of ~2.5% of the genome, including all CpG islands. In terms of both its magnitude and extension along the chromatin, tumour-associated DNA hypermethylation in these regions was much more conspicuous than that observed in the normal mucosal samples from older (vs. younger) tumour-free donors. 83% of the ageing-associated hypermethylated regions (n = 2501) coincided with hypermethylated regions observed in tumour samples. However, 86% of the regions displaying hypermethylation in precancerous lesions (n = 16,772) showed no methylation changes in the ageing normal mucosa. The tumour-specificity of this latter hypermethylation was validated using published sets of data on DNA methylation in normal and neoplastic colon tissues. This extensive set of genomic regions displaying tumour-specific hypermethylation represents a rich vein of putative biomarkers for the early, non-invasive detection of colorectal tumours in women of all ages.
Keywords: DNA methylation; ageing; colorectal adenoma; colorectal cancer; differentially methylated region; normal colorectal mucosa; sessile serrated lesion.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Arndt V, Feller A, Hauri D, et al. Swiss cancer report 2015. Neuchatel, Switzerland: Federal Statistical Office; 2016;
-
- Winawer SJ. Natural history of colorectal cancer. Am J Medicine. 1999;106:3–6. - PubMed
-
- Lash RH, Genta RM, Schuler CM.. Sessile serrated adenomas: prevalence of dysplasia and carcinoma in 2139 patients. J Clin Pathol. 2010;63:681. - PubMed
-
- Hassan C, Quintero E, Dumonceau J-M, et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2013;45:842–864. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
