JWES: a new pipeline for whole genome/exome sequence data processing, management, and gene-variant discovery, annotation, prediction, and genotyping
- PMID: 34370400
- PMCID: PMC8409305
- DOI: 10.1002/2211-5463.13261
JWES: a new pipeline for whole genome/exome sequence data processing, management, and gene-variant discovery, annotation, prediction, and genotyping
Abstract
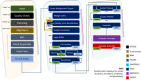
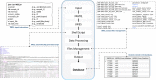
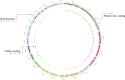
Whole genome and exome sequencing (WGS/WES) are the most popular next-generation sequencing (NGS) methodologies and are at present often used to detect rare and common genetic variants of clinical significance. We emphasize that automated sequence data processing, management, and visualization should be an indispensable component of modern WGS and WES data analysis for sequence assembly, variant detection (SNPs, SVs), imputation, and resolution of haplotypes. In this manuscript, we present a newly developed findable, accessible, interoperable, and reusable (FAIR) bioinformatics-genomics pipeline Java based Whole Genome/Exome Sequence Data Processing Pipeline (JWES) for efficient variant discovery and interpretation, and big data modeling and visualization. JWES is a cross-platform, user-friendly, product line application, that entails three modules: (a) data processing, (b) storage, and (c) visualization. The data processing module performs a series of different tasks for variant calling, the data storage module efficiently manages high-volume gene-variant data, and the data visualization module supports variant data interpretation with Circos graphs. The performance of JWES was tested and validated in-house with different experiments, using Microsoft Windows, macOS Big Sur, and UNIX operating systems. JWES is an open-source and freely available pipeline, allowing scientists to take full advantage of all the computing resources available, without requiring much computer science knowledge. We have successfully applied JWES for processing, management, and gene-variant discovery, annotation, prediction, and genotyping of WGS and WES data to analyze variable complex disorders. In summary, we report the performance of JWES with some reproducible case studies, using open access and in-house generated, high-quality datasets.
Keywords: bioinformatics application; database; gene; variants; whole exome; whole genome.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Meyers BC, Scalabrin S and Morgante M (2004) Mapping and sequencing complex genomes: let's get physical! Nat Rev Genet 5, 578–588. - PubMed
-
- Venter JC, Adams MD, Myers EWet al. (2001) The sequence of the human genome. Science 291, 1304–1351. - PubMed
-
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

