The Corynebacterium diphtheriae HbpA Hemoglobin-Binding Protein Contains a Domain That Is Critical for Hemoprotein Binding, Cellular Localization, and Function
- PMID: 34370560
- PMCID: PMC8508117
- DOI: 10.1128/JB.00196-21
The Corynebacterium diphtheriae HbpA Hemoglobin-Binding Protein Contains a Domain That Is Critical for Hemoprotein Binding, Cellular Localization, and Function
Abstract
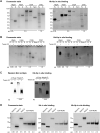
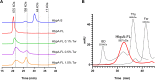
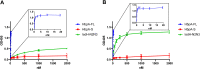
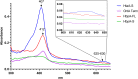
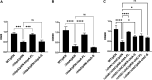
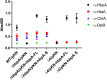
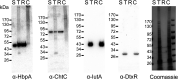
The acquisition of hemin iron from hemoglobin-haptoglobin (Hb-Hp) by Corynebacterium diphtheriae requires the iron-regulated surface proteins HtaA, ChtA, and ChtC and the recently identified Hb-Hp-binding protein, HbpA. We previously showed that a purified form of HbpA (HbpA-S), lacking the C-terminal region, was able to bind Hb-Hp. In this study, we show that the C-terminal region of HbpA significantly enhances binding to Hb-Hp. A purified form of HbpA that includes the C-terminal domain (HbpA-FL) exhibits much stronger binding to Hb-Hp than HbpA-S. Size exclusion chromatography (SEC) showed that HbpA-FL as well as HtaA-FL, ChtA-FL, and ChtC-FL exist as high-molecular-weight complexes, while HbpA-S is present as a monomer, indicating that the C-terminal region is required for formation of large aggregates. Growth studies showed that expression of HbpA-FL in the ΔhbpA mutant restored wild-type levels of growth in low-iron medium that contained Hb-Hp as the sole iron source, while HbpA-S failed to complement the ΔhbpA mutant. Protein localization studies in C. diphtheriae showed that HbpA-FL is present in both the supernatant and membrane fractions and that the C-terminal region is required for membrane anchoring. Purified HbpA-FL was able to enhance growth of the ΔhbpA mutant when added to culture medium that contained Hb-Hp as a sole iron source, suggesting that secreted HbpA is involved in the use of hemin iron from Hb-Hp. These studies extend our understanding of this novel Hb-Hp binding protein in this important human pathogen. IMPORTANCE Hemoproteins, such as Hb, are an abundant source of iron in humans and are proposed to be required by numerous pathogens to cause disease. In this report, we expand on our previous studies in further defining the role of HbpA in hemin iron acquisition in C. diphtheriae. HbpA is unique to C. diphtheriae and appears to function unlike any previously described bacterial iron-regulated Hb- or Hb-Hp-binding protein. HbpA is both secreted and present in the membrane and exists as a large aggregate that enhances its ability to bind Hb-Hp and promote hemin iron uptake. Current studies with HbpA will increase our understanding of iron transport systems in C. diphtheriae.
Keywords: Corynebacterium; diphtheria; hemin; hemoglobin; iron transport.
Figures



References
-
- Zasada AA. 2015. Corynebacterium diphtheriae infections currently and in the past. Przegl Epidemiol 69:439–444, 569-574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

