Transcriptomic and Epigenetic Profiling of Fibroblasts in Idiopathic Pulmonary Fibrosis
- PMID: 34370624
- PMCID: PMC8803355
- DOI: 10.1165/rcmb.2020-0437OC
Transcriptomic and Epigenetic Profiling of Fibroblasts in Idiopathic Pulmonary Fibrosis
Abstract
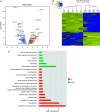
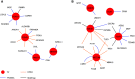
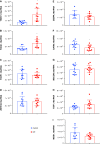
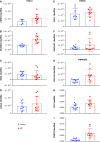
Idiopathic pulmonary fibrosis (IPF), a devastating, fibroproliferative, chronic lung disorder, is associated with expansion of fibroblasts/myofibroblasts, which leads to excessive production and deposition of extracellular matrix. IPF is typically clinically identified as end-stage lung disease, after fibrotic processes are well-established and advanced. Fibroblasts have been shown to be critically important in the development and progression of IPF. We hypothesize that differential chromatin access can drive genetic differences in IPF fibroblasts relative to healthy fibroblasts. To this end, we performed assay of transposase-accessible chromatin sequencing to identify differentially accessible regions within the genomes of fibroblasts from healthy and IPF lungs. Multiple motifs were identified to be enriched in IPF fibroblasts compared with healthy fibroblasts, including binding motifs for TWIST1 and FOXA1. RNA sequencing identified 93 genes that could be annotated to differentially accessible regions. Pathway analysis of the annotated genes identified cellular adhesion, cytoskeletal anchoring, and cell differentiation as important biological processes. In addition, single nucleotide polymorphism analysis showed that linkage disequilibrium blocks of IPF risk single nucleotide polymorphisms with IPF-accessible regions that have been identified to be located in genes that are important in IPF, including MUC5B, TERT, and TOLLIP. Validation studies in isolated lung tissue confirmed increased expression for TWIST1 and FOXA1 in addition to revealing SHANK2 and CSPR2 as novel targets. Thus, modulation of differential chromatin access may be an important mechanism in the pathogenesis of lung fibrosis.
Keywords: ATAC sequencing; RNA sequencing; epigenetics; fibroblasts; idiopathic pulmonary fibrosis.
Figures

Comment in
-
Ready and Waiting: Where Early-Stage Idiopathic Pulmonary Fibrosis Fibroblasts Are Primed to Be Activated.Am J Respir Cell Mol Biol. 2022 Jan;66(1):1-2. doi: 10.1165/rcmb.2021-0365ED. Am J Respir Cell Mol Biol. 2022. PMID: 34533418 Free PMC article. No abstract available.
References
-
- Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med . 2018;378:1811–1823. - PubMed
-
- Hunninghake GW, Lynch DA, Galvin JR, Gross BH, Müller N, Schwartz DA, et al. Radiologic findings are strongly associated with a pathologic diagnosis of usual interstitial pneumonia. Chest . 2003;124:1215–1223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

