Molecular phylogeny of Diplostomum, Tylodelphys, Austrodiplostomum and Paralaria (Digenea: Diplostomidae) necessitates systematic changes and reveals a history of evolutionary host switching events
- PMID: 34371018
- PMCID: PMC8742756
- DOI: 10.1016/j.ijpara.2021.06.002
Molecular phylogeny of Diplostomum, Tylodelphys, Austrodiplostomum and Paralaria (Digenea: Diplostomidae) necessitates systematic changes and reveals a history of evolutionary host switching events
Abstract
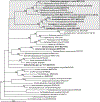
The Diplostomidae Poirier, 1886 is a large, globally distributed family of digeneans parasitic in intestines of their definitive hosts. Diplostomum and Tylodelphys spp. are broadly distributed, commonly reported, and the most often sequenced diplostomid genera. The majority of published DNA sequences from these genera originated from larval stages only, which typically cannot be identified to the species level based on morphology alone. We generated partial large ribosomal subunit (28S) rRNA and cytochrome c oxidase subunit 1 (cox1) mtDNA gene sequences from 14 species/species-level lineages of Diplostomum, six species/species-level lineages of Tylodelphys, two species/species-level lineages of Austrodiplostomum, one species previously assigned to Paralaria, two species/species-level lineages of Dolichorchis and one unknown diplostomid. Our DNA sequences of 11 species/species-level lineages of Diplostomum (all identified to species), four species/species-level lineages of Tylodelphys (all identified to species), Austrodiplostomum compactum, Paralaria alarioides and Dolichorchis lacombeensis originated from adult specimens. 28S sequences were used for phylogenetic inference to demonstrate the position of Paralaria alarioides and Dolichorchis spp. within the Diplostomoidea and study the interrelationships of Diplostomum, Tylodelphys and Austrodiplostomum. Our results demonstrate that two diplostomids from the North American river otter (P. alarioides and a likely undescribed taxon) belong within Diplostomum. Further, our results demonstrate the non-monophyly of Tylodelphys due to the position of Austrodiplostomum spp., based on our phylogenetic analyses and morphology. Furthermore, the results of phylogenetic analysis of 28S confirmed the status of Dolichorchis as a separate genus. The phylogenies suggest multiple definitive host-switching events (birds to otters and among major avian groups) and a New World origin of Diplostomum and Tylodelphys spp. Our DNA sequences from adult digeneans revealed identities of 10 previously published lineages of Diplostomum and Tylodelphys, which were previously identified to genus only. The novel DNA data from this work provide opportunities for future comparisons of larval diplostomines collected in ecological studies.
Keywords: 28S; Cox1; Diplostomidae; Diplostomum; Molecular phylogeny; Tylodelphys.
Copyright © 2021 Australian Society for Parasitology. Published by Elsevier Ltd. All rights reserved.
Figures



References
-
- Achatz TJ, Dmytrieva I, Kuzmin Y, Tkach VV, 2019b. Phylogenetic position of Codonocephalus Diesing, 1850 (Digenea, Diplostomoidea), an unusual diplostomid with progenetic metacercariae. J. Parasitol. 105, 821–826. - PubMed
-
- Achatz TJ, Pulis EE, González-Acuña D, Tkach VV, 2020. Phylogenetic relationships of Cardiocephaloides spp. (Digenea, Diplostomoidea) and the genetic characterization of Cardiocephaloides physalis from Magellanic penguin, Spheniscus magellanicus, in Chile. Acta Parasitol. 65, 525–534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

