The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation
- PMID: 34371659
- PMCID: PMC8309317
- DOI: 10.3390/plants10071456
The Coordinated Upregulated Expression of Genes Involved in MEP, Chlorophyll, Carotenoid and Tocopherol Pathways, Mirrored the Corresponding Metabolite Contents in Rice Leaves during De-Etiolation
Abstract
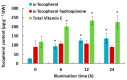
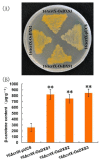
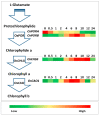
Light is an essential regulator of many developmental processes in higher plants. We investigated the effect of 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 1/2 genes (OsHDR1/2) and isopentenyl diphosphate isomerase 1/2 genes (OsIPPI1/2) on the biosynthesis of chlorophylls, carotenoids, and phytosterols in 14-day-old etiolated rice (Oyza sativa L.) leaves during de-etiolation. However, little is known about the effect of isoprenoid biosynthesis genes on the corresponding metabolites during the de-etiolation of etiolated rice leaves. The results showed that the levels of α-tocopherol were significantly increased in de-etiolated rice leaves. Similar to 1-deoxy-D-xylulose-5-phosphate synthase 3 gene (OsDXS3), both OsDXS1 and OsDXS2 genes encode functional 1-deoxy-D-xylulose-5-phosphate synthase (DXS) activities. Their expression patterns and the synthesis of chlorophyll, carotenoid, and tocopherol metabolites suggested that OsDXS1 is responsible for the biosynthesis of plastidial isoprenoids in de-etiolated rice leaves. The expression analysis of isoprenoid biosynthesis genes revealed that the coordinated expression of the MEP (2-C-methyl-D-erythritol 4-phosphate) pathway, chlorophyll, carotenoid, and tocopherol pathway genes mirrored the changes in the levels of the corresponding metabolites during de-etiolation. The underpinning mechanistic basis of coordinated light-upregulated gene expression was elucidated during the de-etiolation process, specifically the role of light-responsive cis-regulatory motifs in the promoter region of these genes. In silico promoter analysis showed that the light-responsive cis-regulatory elements presented in all the promoter regions of each light-upregulated gene, providing an important link between observed phenotype during de-etiolation and the molecular machinery controlling expression of these genes.
Keywords: carotenoids; coordinated gene expression; isoprenoids; light-upregulated genes; rice (Oryza sativa L.); secondary metabolites.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Chory J., Chatterjee M., Cook R.K., Elich T., Fankhauser C., Li J., Nagpal P., Neff M., Pepper A., Poole D., et al. From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc. Natl. Acad. Sci. USA. 1996;93:12066–12071. doi: 10.1073/pnas.93.22.12066. - DOI - PMC - PubMed
Grants and funding
- 31870278/National Natural Science Foundation of China
- RTI2018-097613-B-I00; PGC2018-097655-B-I00/he Spanish Ministry of Economy and Competitiveness (MINECO), Spain
- 613513/the European Union Framework Program DISCO
- OC-2015-1-19780/the European Cooperation in Science and Technology project EUROCAROTEN
- 2017 SGR 828/Generalitat de Catalunya Grant
LinkOut - more resources
Full Text Sources

