Targeting Viral Surface Proteins through Structure-Based Design
- PMID: 34372526
- PMCID: PMC8310314
- DOI: 10.3390/v13071320
Targeting Viral Surface Proteins through Structure-Based Design
Abstract
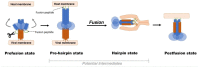
The emergence of novel viral infections of zoonotic origin and mutations of existing human pathogenic viruses represent a serious concern for public health. It warrants the establishment of better interventions and protective therapies to combat the virus and prevent its spread. Surface glycoproteins catalyzing the fusion of viral particles and host cells have proven to be an excellent target for antivirals as well as vaccines. This review focuses on recent advances for computational structure-based design of antivirals and vaccines targeting viral fusion machinery to control seasonal and emerging respiratory viruses.
Keywords: computational protein design; glycoproteins; rational design; respiratory viruses; structural vaccinology; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

