Viroplasms: Assembly and Functions of Rotavirus Replication Factories
- PMID: 34372555
- PMCID: PMC8310052
- DOI: 10.3390/v13071349
Viroplasms: Assembly and Functions of Rotavirus Replication Factories
Abstract
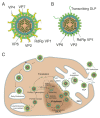
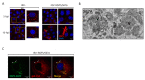
Viroplasms are cytoplasmic, membraneless structures assembled in rotavirus (RV)-infected cells, which are intricately involved in viral replication. Two virus-encoded, non-structural proteins, NSP2 and NSP5, are the main drivers of viroplasm formation. The structures (as far as is known) and functions of these proteins are described. Recent studies using plasmid-only-based reverse genetics have significantly contributed to elucidation of the crucial roles of these proteins in RV replication. Thus, it has been recognized that viroplasms resemble liquid-like protein-RNA condensates that may be formed via liquid-liquid phase separation (LLPS) of NSP2 and NSP5 at the early stages of infection. Interactions between the RNA chaperone NSP2 and the multivalent, intrinsically disordered protein NSP5 result in their condensation (protein droplet formation), which plays a central role in viroplasm assembly. These droplets may provide a unique molecular environment for the establishment of inter-molecular contacts between the RV (+)ssRNA transcripts, followed by their assortment and equimolar packaging. Future efforts to improve our understanding of RV replication and genome assortment in viroplasms should focus on their complex molecular composition, which changes dynamically throughout the RV replication cycle, to support distinct stages of virion assembly.
Keywords: CRISPR-Csy4 genome editing; NSP2; NSP5; liquid-liquid phase separation; protein-RNA condensates; replication cycle; reverse genetics; rotavirus; viroplasm.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Estes M.K., Greenberg H.B. Rotaviruses. In: Knipe D.M., Howley P.M., Cohen J.I., Griffin D.E., Lamb R.A., Martin M.A., Racaniello V.R., Roizman B., editors. Fields Virology. 6th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1347–1401.
-
- Troeger C., Khalil I.A., Rao P.C., Cao S., Blacker B.F., Ahmed T., Armah G., Bines J.E., Brewer T.G., Colombara D.V., et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018;172:958–965. doi: 10.1001/jamapediatrics.2018.1960. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

