Unfolded Protein Response Inhibition Reduces Middle East Respiratory Syndrome Coronavirus-Induced Acute Lung Injury
- PMID: 34372702
- PMCID: PMC8406233
- DOI: 10.1128/mBio.01572-21
Unfolded Protein Response Inhibition Reduces Middle East Respiratory Syndrome Coronavirus-Induced Acute Lung Injury
Abstract
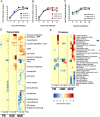
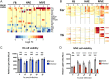
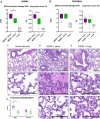
Tissue- and cell-specific expression patterns are highly variable within and across individuals, leading to altered host responses after acute virus infection. Unraveling key tissue-specific response patterns provides novel opportunities for defining fundamental mechanisms of virus-host interaction in disease and the identification of critical tissue-specific networks for disease intervention in the lung. Currently, there are no approved therapeutics for Middle East respiratory syndrome coronavirus (MERS-CoV) patients, and little is understood about how lung cell types contribute to disease outcomes. MERS-CoV replicates equivalently in primary human lung microvascular endothelial cells (MVE) and fibroblasts (FB) and to equivalent peak titers but with slower replication kinetics in human airway epithelial cell cultures (HAE). However, only infected MVE demonstrate observable virus-induced cytopathic effect. To explore mechanisms leading to reduced MVE viability, donor-matched human lung MVE, HAE, and FB were infected, and their transcriptomes, proteomes, and lipidomes were monitored over time. Validated functional enrichment analysis demonstrated that MERS-CoV-infected MVE were dying via an unfolded protein response (UPR)-mediated apoptosis. Pharmacologic manipulation of the UPR in MERS-CoV-infected primary lung cells reduced viral titers and in male mice improved respiratory function with accompanying reductions in weight loss, pathological signatures of acute lung injury, and times to recovery. Systems biology analysis and validation studies of global kinetic transcript, protein, and lipid data sets confirmed that inhibition of host stress pathways that are differentially regulated following MERS-CoV infection of different tissue types can alleviate symptom progression to end-stage lung disease commonly seen following emerging coronavirus outbreaks. IMPORTANCE Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe atypical pneumonia in infected individuals, but the underlying mechanisms of pathogenesis remain unknown. While much has been learned from the few reported autopsy cases, an in-depth understanding of the cells targeted by MERS-CoV in the human lung and their relative contribution to disease outcomes is needed. The host response in MERS-CoV-infected primary human lung microvascular endothelial (MVE) cells and fibroblasts (FB) was evaluated over time by analyzing total RNA, proteins, and lipids to determine the cellular pathways modulated postinfection. Findings revealed that MERS-CoV-infected MVE cells die via apoptotic mechanisms downstream of the unfolded protein response (UPR). Interruption of enzymatic processes within the UPR in MERS-CoV-infected male mice reduced disease symptoms, virus-induced lung injury, and time to recovery. These data suggest that the UPR plays an important role in MERS-CoV infection and may represent a host target for therapeutic intervention.
Keywords: MERS-CoV; acute lung injury; apoptosis; fibroblasts; microvascular endothelial cells; primary human lung cells; unfolded protein response.
Figures


References
-
- Ng DL, Al Hosani F, Keating MK, Gerber SI, Jones TL, Metcalfe MG, Tong S, Tao Y, Alami NN, Haynes LM, Mutei MA, Abdel-Wareth L, Uyeki TM, Swerdlow DL, Barakat M, Zaki SR. 2016. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 186:652–658. doi:10.1016/j.ajpath.2015.10.024. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
