Evaluation of global and intragenic hypomethylation in colorectal adenomas improves patient stratification and colorectal cancer risk prediction
- PMID: 34372923
- PMCID: PMC8351348
- DOI: 10.1186/s13148-021-01135-0
Evaluation of global and intragenic hypomethylation in colorectal adenomas improves patient stratification and colorectal cancer risk prediction
Erratum in
-
Correction to: Evaluation of global and intragenic hypomethylation in colorectal adenomas improves patient stratification and colorectal cancer risk prediction.Clin Epigenetics. 2021 Sep 21;13(1):178. doi: 10.1186/s13148-021-01166-7. Clin Epigenetics. 2021. PMID: 34548107 Free PMC article. No abstract available.
Abstract
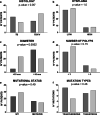
Background: Aberrant DNA hypomethylation of the long interspersed nuclear elements (LINE-1 or L1) has been recognized as an early event of colorectal transformation. Simultaneous genetic and epigenetic analysis of colorectal adenomas may be an effective and rapid strategy to identify key biological features leading to accelerated colorectal tumorigenesis. In particular, global and/or intragenic LINE-1 hypomethylation of adenomas may represent a helpful tool for improving colorectal cancer (CRC) risk stratification of patients after surgical removal of polyps. To verify this hypothesis, we analyzed a cohort of 102 adenomas derived from 40 high-risk patients (who developed CRC in a post-polypectomy of at least one year) and 43 low-risk patients (who did not develop CRC in a post-polypectomy of at least 5 years) for their main pathological features, the presence of hotspot variants in driver oncogenes (KRAS, NRAS, BRAF and PIK3CA), global (LINE-1) and intragenic (L1-MET) methylation status.
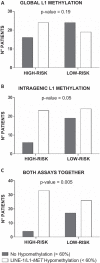
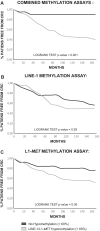
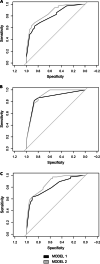
Results: In addition to a significantly higher adenoma size and an older patients' age, adenomas from high-risk patients were more hypomethylated than those from low-risk patients for both global and intragenic LINE-1 assays. DNA hypomethylation, measured by pyrosequencing, was independent from other parameters, including the presence of oncogenic hotspot variants detected by mass spectrometry. Combining LINE-1 and L1-MET analyses and profiling the samples according to the presence of at least one hypomethylated assay improved the discrimination between high and low risk lesions (p = 0.005). Remarkably, adenomas with at least one hypomethylated assay identified the patients with a significantly (p < 0.001) higher risk of developing CRC. Multivariable analysis and logistic regression evaluated by the ROC curves proved that methylation status was an independent variable improving cancer risk prediction (p = 0.02).
Conclusions: LINE-1 and L1-MET hypomethylation in colorectal adenomas are associated with a higher risk of developing CRC. DNA global and intragenic hypomethylation are independent markers that could be used in combination to successfully improve the stratification of patients who enter a colonoscopy surveillance program.
Keywords: Bisulfite pyrosequencing; CRC risk; Colorectal adenomas; L1-MET; LINE-1 hypomethylation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Miglio U, Berrino E, Panero M, Ferrero G, Coscujuela Tarrero L, Miano V, Dell'Aglio C, Sarotto I, Annaratone L, Marchio C, et al. The expression of LINE-1-MET chimeric transcript identifies a subgroup of aggressive breast cancers. Int J Cancer. 2018;143:2838–2848. doi: 10.1002/ijc.31831. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

