Genetic surveillance in the Greater Mekong subregion and South Asia to support malaria control and elimination
- PMID: 34372970
- PMCID: PMC8354633
- DOI: 10.7554/eLife.62997
Genetic surveillance in the Greater Mekong subregion and South Asia to support malaria control and elimination
Abstract
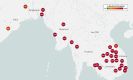
Background: National Malaria Control Programmes (NMCPs) currently make limited use of parasite genetic data. We have developed GenRe-Mekong, a platform for genetic surveillance of malaria in the Greater Mekong Subregion (GMS) that enables NMCPs to implement large-scale surveillance projects by integrating simple sample collection procedures in routine public health procedures.
Methods: Samples from symptomatic patients are processed by SpotMalaria, a high-throughput system that produces a comprehensive set of genotypes comprising several drug resistance markers, species markers and a genomic barcode. GenRe-Mekong delivers Genetic Report Cards, a compendium of genotypes and phenotype predictions used to map prevalence of resistance to multiple drugs.
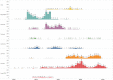
Results: GenRe-Mekong has worked with NMCPs and research projects in eight countries, processing 9623 samples from clinical cases. Monitoring resistance markers has been valuable for tracking the rapid spread of parasites resistant to the dihydroartemisinin-piperaquine combination therapy. In Vietnam and Laos, GenRe-Mekong data have provided novel knowledge about the spread of these resistant strains into previously unaffected provinces, informing decision-making by NMCPs.
Conclusions: GenRe-Mekong provides detailed knowledge about drug resistance at a local level, and facilitates data sharing at a regional level, enabling cross-border resistance monitoring and providing the public health community with valuable insights. The project provides a rich open data resource to benefit the entire malaria community.
Funding: The GenRe-Mekong project is funded by the Bill and Melinda Gates Foundation (OPP11188166, OPP1204268). Genotyping and sequencing were funded by the Wellcome Trust (098051, 206194, 203141, 090770, 204911, 106698/B/14/Z) and Medical Research Council (G0600718). A proportion of samples were collected with the support of the UK Department for International Development (201900, M006212), and Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Keywords: asia; drug resistance; epidemiology; genetic surveillance; global health; infectious disease; malaria; microbiology.
Conflict of interest statement
CJ, NT, MM, RM, HQ, BH, VV, TN, HR, Rv, Lv, RF, FN, MH, NP, SG, PR, KC, PN, EA, SP, RM, RL, CH, LD, KN, TN, TH, AS, DU, CB, CF, MO, TP, RT, CA, AM, GD, JL, DP, NC, DL, SS, JC, PJ, BH, SP, AP, FS, KL, MT, TH, PS, SS, PB, AT, SB, NV, AA, AU, AF, CK, ED, MK, MA, KL, SR, AJ, KR, CH, MD, RV, NK, PW, JA, RP, CA, TC, CM, TN, JS, BJ, JK, KJ, DM, XC, JS, RA, VS, SG, KR, ND, AD, DK, OM No competing interests declared, HN, NZ, SC is an employee of Vysnova Partners Inc
Figures














Comment in
-
A genetic intervention.Elife. 2021 Aug 12;10:e72000. doi: 10.7554/eLife.72000. Elife. 2021. PMID: 34382938 Free PMC article.
References
-
- Agrawal S, Moser KA, Morton L, Cummings MP, Parihar A, Dwivedi A, Shetty AC, Drabek EF, Jacob CG, Henrich PP, Parobek CM, Jongsakul K, Huy R, Spring MD, Lanteri CA, Chaorattanakawee S, Lon C, Fukuda MM, Saunders DL, Fidock DA, Lin JT, Juliano JJ, Plowe CV, Silva JC, Takala-Harrison S. Association of a novel mutation in the Plasmodium falciparum chloroquine resistance transporter with decreased piperaquine sensitivity. The Journal of Infectious Diseases. 2017;216:468–476. doi: 10.1093/infdis/jix334. - DOI - PMC - PubMed
-
- Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. Dihydroartemisinin–piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. The Lancet Infectious Diseases. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. - DOI - PMC - PubMed
-
- Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. The Lancet Infectious Diseases. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. - DOI - PMC - PubMed
-
- Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, Fairhurst RM, Kwiatkowski DP. Origins of the current outbreak of multidrug-resistant malaria in Southeast Asia: a retrospective genetic study. The Lancet Infectious Diseases. 2018;18:337–345. doi: 10.1016/S1473-3099(18)30068-9. - DOI - PMC - PubMed
-
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

