Efficient integration of transmembrane domains depends on the folding properties of the upstream sequences
- PMID: 34373330
- PMCID: PMC8379923
- DOI: 10.1073/pnas.2102675118
Efficient integration of transmembrane domains depends on the folding properties of the upstream sequences
Abstract
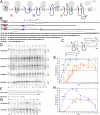
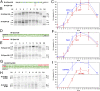
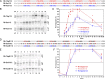
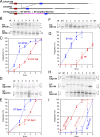
The topology of most membrane proteins is defined by the successive integration of α-helical transmembrane domains at the Sec61 translocon. The translocon provides a pore for the transfer of polypeptide segments across the membrane while giving them lateral access to the lipid. For each polypeptide segment of ∼20 residues, the combined hydrophobicities of its constituent amino acids were previously shown to define the extent of membrane integration. Here, we discovered that different sequences preceding a potential transmembrane domain substantially affect its hydrophobicity requirement for integration. Rapidly folding domains, sequences that are intrinsically disordered or very short or capable of binding chaperones with high affinity, allow for efficient transmembrane integration with low-hydrophobicity thresholds for both orientations in the membrane. In contrast, long protein fragments, folding-deficient mutant domains, and artificial sequences not binding chaperones interfered with membrane integration, requiring higher hydrophobicity. We propose that the latter sequences, as they compact on their hydrophobic residues, partially folded but unable to reach a native state, expose hydrophobic surfaces that compete with the translocon for the emerging transmembrane segment, reducing integration efficiency. The results suggest that rapid folding or strong chaperone binding is required for efficient transmembrane integration.
Keywords: Sec61 translocon; membrane proteins; molecular chaperones; protein folding; topogenesis.
Copyright 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


Comment in
-
Molten globules lure transmembrane helices away from the membrane.Proc Natl Acad Sci U S A. 2021 Aug 31;118(35):e2112899118. doi: 10.1073/pnas.2112899118. Proc Natl Acad Sci U S A. 2021. PMID: 34433673 Free PMC article. No abstract available.
References
-
- Spiess M., Junne T., Janoschke M., Membrane protein integration and topogenesis at the ER. Protein J. 38, 306–316 (2019). - PubMed
-
- Rapoport T. A., Li L., Park E., Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 33, 369–390 (2017). - PubMed
-
- Gogala M., et al. ., Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

