Integrated single-cell analysis revealed immune dynamics during Ad5-nCoV immunization
- PMID: 34373443
- PMCID: PMC8352953
- DOI: 10.1038/s41421-021-00300-2
Integrated single-cell analysis revealed immune dynamics during Ad5-nCoV immunization
Abstract
Coronavirus disease 2019 (COVID-19), driven by SARS-CoV-2, is a severe infectious disease that has become a global health threat. Vaccines are among the most effective public health tools for combating COVID-19. Immune status is critical for evaluating the safety and response to the vaccine, however, the evolution of the immune response during immunization remains poorly understood. Single-cell RNA sequencing (scRNA-seq) represents a powerful tool for dissecting multicellular behavior and discovering therapeutic antibodies. Herein, by performing scRNA/V(D)J-seq on peripheral blood mononuclear cells from four COVID-19 vaccine trial participants longitudinally during immunization, we revealed enhanced cellular immunity with concerted and cell type-specific IFN responses as well as boosted humoral immunity with SARS-CoV-2-specific antibodies. Based on the CDR3 sequence and germline enrichment, we were able to identify several potential binding antibodies. We synthesized, expressed and tested 21 clones from the identified lineages. Among them, one monoclonal antibody (P3V6-1) exhibited relatively high affinity with the extracellular domain of Spike protein, which might be a promising therapeutic reagent for COVID-19. Overall, our findings provide insights for assessing vaccine through the novel scRNA/V(D)J-seq approach, which might facilitate the development of more potent, durable and safe prophylactic vaccines.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


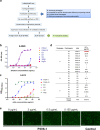


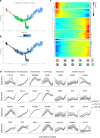
References
-
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU), https://coronavirus.jhu.edu/map.html (2021).
Grants and funding
- 81722034/National Natural Science Foundation of China (National Science Foundation of China)
- 81988101/National Natural Science Foundation of China (National Science Foundation of China)
- 81802878/National Natural Science Foundation of China (National Science Foundation of China)
- 81670015/National Natural Science Foundation of China (National Science Foundation of China)
- 2018ZX09101002/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)
LinkOut - more resources
Full Text Sources
Miscellaneous

