CREB signaling activity correlates with differentiation and survival in medulloblastoma
- PMID: 34373489
- PMCID: PMC8352923
- DOI: 10.1038/s41598-021-95381-0
CREB signaling activity correlates with differentiation and survival in medulloblastoma
Abstract
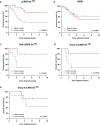
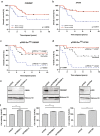
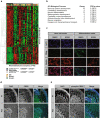
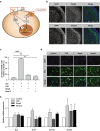
While there has been significant progress in the molecular characterization of the childhood brain cancer medulloblastoma, the tumor proteome remains less explored. However, it is important to obtain a complete understanding of medulloblastoma protein biology, since interactions between proteins represent potential new drug targets. Using previously generated phosphoprotein signaling-profiles of a large cohort of primary medulloblastoma, we discovered that phosphorylation of transcription factor CREB strongly correlates with medulloblastoma survival and associates with a differentiation phenotype. We further found that during normal cerebellar development, phosphorylated CREB was selectively expressed in differentiating cerebellar granule neuron progenitor (CGNP) cells. In line, we observed increased differentiation in CGNPs treated with Forskolin, Bmp6 and Bmp12 (Gdf7), which induce CREB phosphorylation. Lastly, we demonstrated that inducing CREB activation via PKA-mediated CREB signaling, but not Bmp/MEK/ERK mediated signalling, enhances medulloblastoma cell sensitivity to chemotherapy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

