High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement
- PMID: 34373642
- PMCID: PMC8423627
- DOI: 10.1038/s41588-021-00910-2
High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement
Abstract
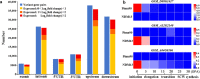
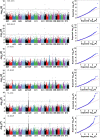
Cotton produces natural fiber for the textile industry. The genetic effects of genomic structural variations underlying agronomic traits remain unclear. Here, we generate two high-quality genomes of Gossypium hirsutum cv. NDM8 and Gossypium barbadense acc. Pima90, and identify large-scale structural variations in the two species and 1,081 G. hirsutum accessions. The density of structural variations is higher in the D-subgenome than in the A-subgenome, indicating that the D-subgenome undergoes stronger selection during species formation and variety development. Many structural variations in genes and/or regulatory regions potentially influencing agronomic traits were discovered. Of 446 significantly associated structural variations, those for fiber quality and Verticillium wilt resistance are located mainly in the D-subgenome and those for yield mainly in the A-subgenome. Our research provides insight into the role of structural variations in genotype-to-phenotype relationships and their potential utility in crop improvement.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science361, eaar 7191 (2018). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

