Genome-wide methylation analysis reveals differentially methylated CpG sites and altered expression of heart development-associated genes in fetuses with cardiac defects
- PMID: 34373718
- PMCID: PMC8343574
- DOI: 10.3892/etm.2021.10464
Genome-wide methylation analysis reveals differentially methylated CpG sites and altered expression of heart development-associated genes in fetuses with cardiac defects
Abstract
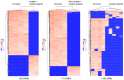
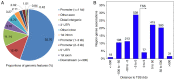
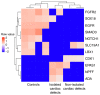
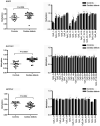
DNA methylation, as an epigenetic mechanism, has a vital role in heart development. An increasing number of studies have investigated aberrant DNA methylation in pediatric or adult heart samples from patients with congenital heart defects (CHD). Placenta tissue, umbilical cord blood, or newborn blood have also been used to detect DNA methylation biomarkers for CHD. However, few studies have compared the methylation levels in fetal heart tissue with cardiac defects with that in normal controls. The present study conducted an integrative whole-genome and CpG site-specific DNA methylation analysis of fetal heart samples from 17 isolated cardiac defect cases, 14 non-isolated cardiac defect cases, and 22 controls with normal hearts, using methylated DNA immunoprecipitation microarray and MassARRAY EpiTYPER assays. Expression of genes adjacent to differentially methylated regions (DMRs) was measured by RT-qPCR and western blot analysis. The results revealed that fetuses with cardiac defects presented global hypomethylation. Genomic analysis of DMRs revealed that a proportion of DMRs were located in exons (12.4%), distal intergenic regions (11.14%), and introns (8.97%). Only 55.7% of DMRs were observed at promoter regions. Functional enrichment analysis for genes adjacent to these DMRs revealed that hypomethylated genes were involved in embryonic heart tube morphogenesis and immune-related regulation functions. Intergenic hypermethylation of EGFR and solute carrier family 19 member 1 (SLC19A1), and intragenic hypomethylation of NOTCH1 were validated in fetal heart tissues with cardiac defects. Only SLC19A1 expression was significantly decreased at the mRNA level, while EGFR, NOTCH1, and SLC19A1 expression were all significantly decreased at the protein level. In conclusion, the present study demonstrated that fetal cardiac defects may be associated with alterations in regional and single CpG site methylation outside of promoter regions, resulting in differentiated expression of corresponding genes associated with heart development. These results present new insights into the epigenetic mechanisms underlying abnormal heart development.
Keywords: CpG site methylation; cardiac defects; fetal; genes expression alterations.
Copyright: © Zhou et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
