Ligase 1 is a predictor of platinum resistance and its blockade is synthetically lethal in XRCC1 deficient epithelial ovarian cancers
- PMID: 34373746
- PMCID: PMC8344016
- DOI: 10.7150/thno.51456
Ligase 1 is a predictor of platinum resistance and its blockade is synthetically lethal in XRCC1 deficient epithelial ovarian cancers
Abstract
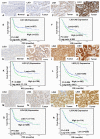
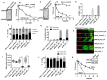
Rationale: The human ligases (LIG1, LIG3 and LIG4) are essential for the maintenance of genomic integrity by catalysing the formation of phosphodiester bonds between adjacent 5'-phosphoryl and 3'-hydroxyl termini at single and double strand breaks in duplex DNA molecules generated either directly by DNA damage or during replication, recombination, and DNA repair. Whether LIG1, LIG3 and LIG4 can influence ovarian cancer pathogenesis and therapeutics is largely unknown. Methods: We investigated LIG1, LIG3 and LIG4 expression in clinical cohorts of epithelial ovarian cancers [protein level (n=525) and transcriptional level (n=1075)] and correlated to clinicopathological features and survival outcomes. Pre-clinically, platinum sensitivity was investigated in LIG1 depleted ovarian cancer cells. A small molecule inhibitor of LIG1 (L82) was tested for synthetic lethality application in XRCC1, BRCA2 or ATM deficient cancer cells. Results: LIG1 and LIG3 overexpression linked with aggressive phenotypes, platinum resistance and poor progression free survival (PFS). In contrast, LIG4 deficiency was associated with platinum resistance and worse PFS. In a multivariate analysis, LIG1 was independently associated with adverse outcome. In ovarian cancer cell lines, LIG1 depletion increased platinum cytotoxicity. L82 monotherapy was synthetically lethal in XRCC1 deficient ovarian cancer cells and 3D-spheroids. Increased cytotoxicity was linked with accumulation of DNA double strand breaks (DSBs), S-phase cell cycle arrest and increased apoptotic cells. L82 was also selectively toxic in BRCA2 deficient or ATM deficient cancer cells and 3D-spheroids. Conclusions: We provide evidence that LIG1 is an attractive target for personalization of ovarian cancer therapy.
Keywords: DNA repair; LIG1; LIG1 inhibitor; LIG3; LIG4; Ovarian cancer; Predictive bimarker; Prognostics; Synthetic lethality.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Konstantinopoulos PA, Lheureux S, Moore KN. PARP Inhibitors for Ovarian Cancer: Current Indications, Future Combinations, and Novel Assets in Development to Target DNA Damage Repair. Am Soc Clin Oncol Educ Book. 2020;40:1–16. - PubMed
-
- Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–70. - PubMed
-
- D'Andrea AD. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst) 2018;71:172–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

